8CBQ
 
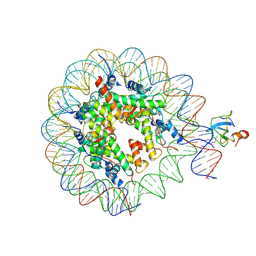 | | structure of LEDGF/p75 PWWP domain bound to the H3K36 trimethylated dinucleosome | | Descriptor: | Histone H2A, Histone H2B 1.1, Histone H3, ... | | Authors: | Koutna, E, Kouba, T, Novacek, J, Veverka, V. | | Deposit date: | 2023-01-25 | | Release date: | 2023-09-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Multivalency of nucleosome recognition by LEDGF.
Nucleic Acids Res., 51, 2023
|
|
6CM3
 
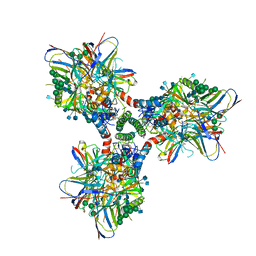 | | BG505 SOSIP in complex with sCD4, 17b, 8ANC195 | | Descriptor: | 17b Fab heavy chain, 17b Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, H, Bjorkman, P.J. | | Deposit date: | 2018-03-02 | | Release date: | 2018-10-17 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Partially Open HIV-1 Envelope Structures Exhibit Conformational Changes Relevant for Coreceptor Binding and Fusion.
Cell Host Microbe, 24, 2018
|
|
8RUS
 
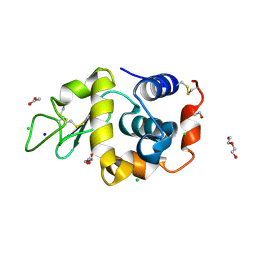 | | Hen egg-white lysozyme (HEWL) structure from EuXFEL FXE, multi-hit Droplet-on-Demand (DoD) injection, 9.3 keV photon energy, space group P432121 | | Descriptor: | ACETATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Perrett, S, van Thor, J.J. | | Deposit date: | 2024-01-31 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Kilohertz droplet-on-demand serial femtosecond crystallography at the European XFEL station FXE.
Struct Dyn., 11, 2024
|
|
8C5U
 
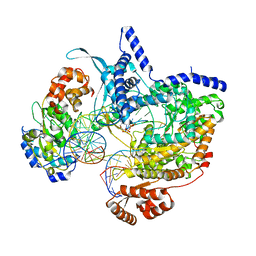 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with 8-mer RNA, pppGpGpUpApApApUpG (IC8) | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2023-01-10 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
6CNM
 
 | | Cryo-EM structure of the human SK4/calmodulin channel complex | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Calmodulin-1, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Lee, C.H, MacKinnon, R. | | Deposit date: | 2018-03-08 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Activation mechanism of a human SK-calmodulin channel complex elucidated by cryo-EM structures.
Science, 360, 2018
|
|
7T3O
 
 | |
6CO1
 
 | |
6JNF
 
 | | Cryo-EM structure of the translocator of the outer mitochondrial membrane | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | Authors: | Araiso, Y, Tsutsumi, A, Suzuki, J, Yunoki, K, Kawano, S, Kikkawa, M, Endo, T. | | Deposit date: | 2019-03-14 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structure of the mitochondrial import gate reveals distinct preprotein paths.
Nature, 575, 2019
|
|
6CB1
 
 | | Yeast nucleolar pre-60S ribosomal subunit (state 3) | | Descriptor: | 35S pre-ribosomal RNA miscRNA, 5.8S rRNA, 60S ribosomal protein L13-A, ... | | Authors: | Sanghai, Z.A, Miller, L, Barandun, J, Hunziker, M, Chaker-Margot, M, Klinge, S. | | Deposit date: | 2018-02-01 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Modular assembly of the nucleolar pre-60S ribosomal subunit.
Nature, 556, 2018
|
|
6COZ
 
 | |
8CC3
 
 | | Vibrio cholerae GbpA (LPMO domain) | | Descriptor: | ACETATE ION, COPPER (II) ION, GlcNAc-binding protein A, ... | | Authors: | Montserrat-Canals, M, Sorensen, H.V, Cordara, G, Krengel, U. | | Deposit date: | 2023-01-26 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.128 Å) | | Cite: | Perdeuterated GbpA Enables Neutron Scattering Experiments of a Lytic Polysaccharide Monooxygenase.
Acs Omega, 8, 2023
|
|
6CRF
 
 | | Crystal Structure of Shp2 E76K GOF Mutant in the Open Conformation | | Descriptor: | GLYCEROL, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Stams, T, Fodor, M. | | Deposit date: | 2018-03-17 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural reorganization of SHP2 by oncogenic mutations and implications for oncoprotein resistance to allosteric inhibition.
Nat Commun, 9, 2018
|
|
6JLF
 
 | |
6CRU
 
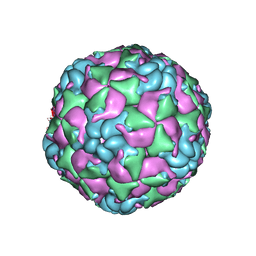 | |
6CS4
 
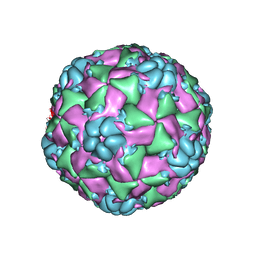 | |
8BYG
 
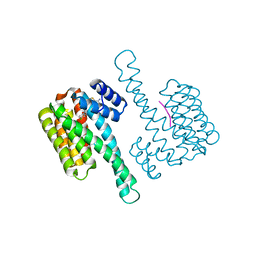 | | fragment-linked stabilizer for ERa - 14-3-3 interaction (1047648) | | Descriptor: | 14-3-3 protein sigma, ERalpha peptide, ~{N}-[2-[(2-carbamimidoyl-1-benzothiophen-4-yl)-methyl-amino]ethyl]-2-(4-chloranylphenoxy)-~{N},2-dimethyl-propanamide | | Authors: | Visser, E.J, Sijbesma, E, Ottmann, C. | | Deposit date: | 2022-12-12 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | From Tethered to Freestanding Stabilizers of 14-3-3 Protein-Protein Interactions through Fragment Linking.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6CS6
 
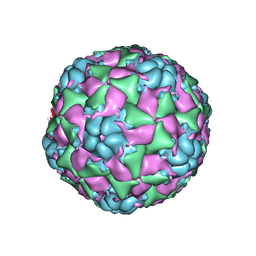 | |
8C4Y
 
 | | SFX structure of FutA bound to Fe(III) | | Descriptor: | FE (III) ION, Putative iron ABC transporter, substrate binding protein | | Authors: | Bolton, R, Tews, I. | | Deposit date: | 2023-01-05 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A redox switch allows binding of Fe(II) and Fe(III) ions in the cyanobacterial iron-binding protein FutA from Prochlorococcus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8BZ9
 
 | | single soak stabilizer for ERa - 14-3-3 interaction (AZ354) | | Descriptor: | 14-3-3 protein sigma, 4-chloranyl-7-propan-2-yloxy-1-benzothiophene-2-carboximidamide, ERalpha peptide, ... | | Authors: | Visser, E.J, Sijbesma, E, Ottmann, C. | | Deposit date: | 2022-12-14 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | From Tethered to Freestanding Stabilizers of 14-3-3 Protein-Protein Interactions through Fragment Linking.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6JSI
 
 | | Co-purified Fatty Acid Synthase | | Descriptor: | Fatty acid synthase subunit alpha, Fatty acid synthase subunit beta | | Authors: | Qiu, S.W, Liu, S. | | Deposit date: | 2019-04-08 | | Release date: | 2019-09-25 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Modulation of fatty acid synthase by ATR checkpoint kinase Rad3.
J Mol Cell Biol, 11, 2019
|
|
8C5S
 
 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with 7-mer RNA, pppGpGpUpApApApU (IC7) | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2023-01-10 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
8CC5
 
 | | Vibrio cholerae GbpA (LPMO domain) | | Descriptor: | ACETATE ION, COPPER (II) ION, GlcNAc-binding protein A, ... | | Authors: | Montserrat-Canals, M, Sorensen, H.V, Cordara, G, Krengel, U. | | Deposit date: | 2023-01-26 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Perdeuterated GbpA Enables Neutron Scattering Experiments of a Lytic Polysaccharide Monooxygenase.
Acs Omega, 8, 2023
|
|
6JN9
 
 | | Cryo-EM structure of glutamate dehydrogenase from Thermococcus profundus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutamate dehydrogenase | | Authors: | Oide, M, Kato, T, Oroguchi, T, Nakasako, M. | | Deposit date: | 2019-03-14 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Energy landscape of domain motion in glutamate dehydrogenase deduced from cryo-electron microscopy.
Febs J., 287, 2020
|
|
8CKP
 
 | | X-ray structure of the crystallization-prone form of subfamily III haloalkane dehalogenase DhmeA from Haloferax mediterranei | | Descriptor: | Alpha/beta fold hydrolase, CHLORIDE ION | | Authors: | Marek, M, Chmelova, K, Schenkmayerova, A, Croll, T, Read, R.J, Diederichs, K. | | Deposit date: | 2023-02-16 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Multimeric structure of a subfamily III haloalkane dehalogenase-like enzyme solved by combination of cryo-EM and x-ray crystallography.
Protein Sci., 32, 2023
|
|
6JNC
 
 | | Cryo-EM structure of glutamate dehydrogenase from Thermococcus profundus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutamate dehydrogenase | | Authors: | Oide, M, Kato, T, Oroguchi, T, Nakasako, M. | | Deposit date: | 2019-03-14 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Energy landscape of domain motion in glutamate dehydrogenase deduced from cryo-electron microscopy.
Febs J., 287, 2020
|
|
