8H2F
 
 | |
8H18
 
 | |
3G6T
 
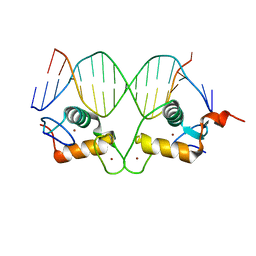 | | GR gamma DNA-binding domain:FKBP5 16bp complex-34 | | Descriptor: | DNA (5'-D(*AP*AP*GP*AP*AP*CP*AP*GP*GP*GP*TP*GP*TP*TP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*AP*AP*CP*AP*CP*CP*CP*TP*GP*TP*TP*CP*T)-3'), Glucocorticoid receptor, ... | | Authors: | Pufall, M.A, Yamamoto, K.R, Meijsing, S.H. | | Deposit date: | 2009-02-08 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DNA binding site sequence directs glucocorticoid receptor structure and activity.
Science, 324, 2009
|
|
6GPI
 
 | | Tc-DNA/DNA duplex | | Descriptor: | DNA (5'-D(*GP*TP*AP*AP*GP*CP*CP*GP*AP*G)-3'), tc-DNA (5'-D(*(TCJ)P*(TTK)P*(TCJ)P*(TCS)P*(TCS)P*(TCJ)P*(TTK)P*(TTK)P*(TCY)P*(TCJ))-3') | | Authors: | Istrate, A, Johannsen, S, Istrate, A, Sigel, R.K.O, Leumann, C. | | Deposit date: | 2018-06-05 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of tricyclo-DNA containing duplexes: insight into enhanced thermal stability and nuclease resistance.
Nucleic Acids Res., 47, 2019
|
|
2BR0
 
 | | DNA Adduct Bypass Polymerization by Sulfolobus solfataricus Dpo4. Analysis and Crystal Structures of Multiple Base-Pair Substitution and Frameshift Products with the Adduct 1,N2-Ethenoguanine | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*C)-3', 5'-D(*TP*CP*AP*CP*GNEP*GP*AP*AP*TP*CP*CP *TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Irimia, A, Loukachevitch, L.V, Egli, M. | | Deposit date: | 2005-04-28 | | Release date: | 2005-06-23 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | DNA Adduct Bypass Polymerization by Sulfolobus Solfataricus DNA Polymerase Dpo4: Analysis and Crystal Structures of Multiple Base Pair Substitution and Frameshift Products with the Adduct 1,N2-Ethenoguanine.
J.Biol.Chem., 280, 2005
|
|
2Z4R
 
 | | Crystal structure of domain III from the Thermotoga maritima replication initiation protein DnaA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromosomal replication initiator protein dnaA, MAGNESIUM ION | | Authors: | Fujikawa, N, Ozaki, S, Kagawa, W, Park, S.-Y, Katayama, T, Kurumizaka, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-06-25 | | Release date: | 2008-02-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | A Common Mechanism for the ATP-DnaA-dependent Formation of Open Complexes at the Replication Origin
J.Biol.Chem., 283, 2008
|
|
2BQU
 
 | | DNA Adduct Bypass Polymerization by Sulfolobus solfataricus Dpo4. Analysis and Crystal Structures of Multiple Base-Pair Substitution and Frameshift Products with the Adduct 1,N2-Ethenoguanine | | Descriptor: | 2',3'-DIDEOXYADENOSINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*C)-3', 5'-D(*TP*CP*AP*TP*GNEP*GP*AP*AP*TP*CP*CP *TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Irimia, A, Loukachevitch, L.V, Egli, M. | | Deposit date: | 2005-04-28 | | Release date: | 2005-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA Adduct Bypass Polymerization by Sulfolobus Solfataricus DNA Polymerase Dpo4: Analysis and Crystal Structures of Multiple Base Pair Substitution and Frameshift Products with the Adduct 1,N2-Ethenoguanine.
J.Biol.Chem., 280, 2005
|
|
2BQR
 
 | | DNA Adduct Bypass Polymerization by Sulfolobus solfataricus Dpo4. Analysis and Crystal Structures of Multiple Base-Pair Substitution and Frameshift Products with the Adduct 1,N2-Ethenoguanine | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*C)-3', 5'-D(*TP*CP*AP*TP*GNEP*GP*AP*AP*TP*CP*CP *TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Irimia, A, Loukachevitch, L.V, Egli, M. | | Deposit date: | 2005-04-27 | | Release date: | 2005-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | DNA Adduct Bypass Polymerization by Sulfolobus Solfataricus DNA Polymerase Dpo4: Analysis and Crystal Structures of Multiple Base Pair Substitution and Frameshift Products with the Adduct 1,N2-Ethenoguanine.
J.Biol.Chem., 280, 2005
|
|
2BQ3
 
 | | DNA Adduct Bypass Polymerization by Sulfolobus solfataricus Dpo4. Analysis and Crystal Structures of Multiple Base-Pair Substitution and Frameshift Products with the Adduct 1,N2-Ethenoguanine | | Descriptor: | 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*CP*A)-3', 5'-D(*TP*CP*AP*TP*GNEP*GP*AP*AP*TP*CP*CP *TP*TP*CP*CP*CP*CP*C)-3', CALCIUM ION, ... | | Authors: | Irimia, A, Loukachevitch, L.V, Egli, M. | | Deposit date: | 2005-04-27 | | Release date: | 2005-06-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA Adduct Bypass Polymerization by Sulfolobus Solfataricus DNA Polymerase Dpo4: Analysis and Crystal Structures of Multiple Base Pair Substitution and Frameshift Products with the Adduct 1,N2-Ethenoguanine.
J.Biol.Chem., 280, 2005
|
|
6QEM
 
 | | E. coli DnaBC complex bound to ssDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA replication protein DnaC, MAGNESIUM ION, ... | | Authors: | Arias-Palomo, E, Puri, N, O'Shea Murray, V.L, Yan, Q, Berger, J.M. | | Deposit date: | 2019-01-08 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Physical Basis for the Loading of a Bacterial Replicative Helicase onto DNA.
Mol.Cell, 74, 2019
|
|
5YV2
 
 | | DNA polymerase IV - DNA ternary complex 14 | | Descriptor: | DNA polymerase IV, DTN1, DTN2, ... | | Authors: | Kottur, J, Nair, D.T. | | Deposit date: | 2017-11-23 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pyrophosphate hydrolysis is an intrinsic and critical step of the DNA synthesis reaction
Nucleic Acids Res., 46, 2018
|
|
5YUW
 
 | | DNA polymerase IV - DNA ternary complex 6 | | Descriptor: | DNA polymerase IV, DTN1, DTN2C, ... | | Authors: | Kottur, J, Nair, D.T. | | Deposit date: | 2017-11-23 | | Release date: | 2018-11-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.124 Å) | | Cite: | Pyrophosphate hydrolysis is an intrinsic and critical step of the DNA synthesis reaction
Nucleic Acids Res., 46, 2018
|
|
3G6Q
 
 | | GR DNA binding domain:FKBP5 binding site complex-9 | | Descriptor: | DNA (5'-D(*AP*AP*GP*AP*AP*CP*AP*CP*CP*CP*TP*GP*TP*TP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*AP*AP*CP*AP*GP*GP*GP*TP*GP*TP*TP*CP*T)-3'), Glucocorticoid receptor, ... | | Authors: | Pufall, M.A, Yamamoto, K.R, Meijsing, S.H. | | Deposit date: | 2009-02-08 | | Release date: | 2009-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | DNA binding site sequence directs glucocorticoid receptor structure and activity.
Science, 324, 2009
|
|
3G9P
 
 | | GR DNA binding domain:Sgk 16bp complex-7 | | Descriptor: | DNA (5'-D(*AP*AP*GP*AP*AP*CP*AP*TP*TP*TP*TP*GP*TP*CP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*GP*AP*CP*AP*AP*AP*AP*TP*GP*TP*TP*CP*T)-3'), Glucocorticoid receptor, ... | | Authors: | Pufall, M.A, Yamamoto, K.R, Meijsing, S.H. | | Deposit date: | 2009-02-13 | | Release date: | 2009-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | DNA binding site sequence directs glucocorticoid receptor structure and activity.
Science, 324, 2009
|
|
3G6R
 
 | | GR DNA binding domain:FKBP5 complex-52, 18bp | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*CP*CP*CP*TP*GP*TP*TP*CP*TP*G)-3'), DNA (5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*GP*GP*GP*TP*GP*TP*TP*CP*TP*G)-3'), Glucocorticoid receptor, ... | | Authors: | Pufall, M.A, Yamamoto, K.R, Meijsing, S.H. | | Deposit date: | 2009-02-08 | | Release date: | 2009-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA binding site sequence directs glucocorticoid receptor structure and activity.
Science, 324, 2009
|
|
6CBS
 
 | |
6CBT
 
 | |
6KNC
 
 | | PolD-PCNA-DNA (form B) | | Descriptor: | DNA polymerase D DP2 (DNA polymerase II large) subunit, DNA polymerase II small subunit, DNA polymerase sliding clamp 1, ... | | Authors: | Mayanagi, K, Oki, K, Miyazaki, N, Ishino, S, Yamagami, T, Iwasaki, K, Kohda, D, Morikawa, K, Shirai, T, Ishino, Y. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Two conformations of DNA polymerase D-PCNA-DNA, an archaeal replisome complex, revealed by cryo-electron microscopy.
Bmc Biol., 18, 2020
|
|
6KNB
 
 | | PolD-PCNA-DNA (form A) | | Descriptor: | DNA polymerase D DP2 (DNA polymerase II large) subunit, DNA polymerase II small subunit, DNA polymerase sliding clamp 1, ... | | Authors: | Mayanagi, K, Oki, K, Miyazaki, N, Ishino, S, Yamagami, T, Iwasaki, K, Kohda, D, Morikawa, K, Shirai, T, Ishino, Y. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-05 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Two conformations of DNA polymerase D-PCNA-DNA, an archaeal replisome complex, revealed by cryo-electron microscopy.
Bmc Biol., 18, 2020
|
|
9FFF
 
 | | dsDNA-FANCD2-FANCI complex | | Descriptor: | DNA (32-MER), DNA (33-MER), Fanconi anemia complementation group I, ... | | Authors: | Alcon, P, Passmore, L.A. | | Deposit date: | 2024-05-23 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | FANCD2-FANCI surveys DNA and recognizes double- to single-stranded junctions.
Nature, 632, 2024
|
|
9FFB
 
 | | ss-dsDNA-FANCD2-FANCI complex | | Descriptor: | DNA (5'-D(P*CP*GP*AP*TP*GP*TP*CP*TP*CP*TP*AP*GP*AP*CP*AP*GP*CP*TP*GP*C)-3'), DNA (5'-D(P*GP*CP*AP*GP*CP*TP*GP*TP*CP*TP*AP*GP*AP*GP*AP*CP*AP*TP*CP*GP*AP*T)-3'), Fanconi anemia complementation group I, ... | | Authors: | Alcon, P, Passmore, L.A. | | Deposit date: | 2024-05-22 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | FANCD2-FANCI surveys DNA and recognizes double- to single-stranded junctions.
Nature, 632, 2024
|
|
4YKL
 
 | | Hnt3 in complex with DNA and guanosine | | Descriptor: | Aprataxin-like protein, CHLORIDE ION, DNA (5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3'), ... | | Authors: | Jacewicz, A, Chauleau, M, Shuman, S. | | Deposit date: | 2015-03-04 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | DNA3'pp5'G de-capping activity of aprataxin: effect of cap nucleoside analogs and structural basis for guanosine recognition.
Nucleic Acids Res., 43, 2015
|
|
8QPC
 
 | | 18mer DNA mimic Foldamer with an Aromatic linker in complex with Sac7d V26A/M29A protein | | Descriptor: | DNA-binding protein 7b, N-[2-(2-methyl-1,3-dioxolan-2-yl)phenyl]-2-{[5-(trifluoromethyl)pyridin-2-yl]amino}pyridine-4-carboxamide | | Authors: | Deepak, D, Corvaglia, V, Wu, J, Huc, I. | | Deposit date: | 2023-10-01 | | Release date: | 2023-11-08 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | DNA Mimic Foldamer Recognition of a Chromosomal Protein.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
8TLQ
 
 | | Cryo-EM structure of the Rev1-Polzeta-DNA-dCTP complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (30-MER), ... | | Authors: | Malik, R, Aggarwal, A.K. | | Deposit date: | 2023-07-27 | | Release date: | 2024-05-15 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Cryo-EM structure of the Rev1-Pol zeta holocomplex reveals the mechanism of their cooperativity in translesion DNA synthesis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
2WP0
 
 | | Crystal structure of a HobA-DnaA (domain I-II) complex from Helicobacter pylori. | | Descriptor: | ACETATE ION, CHLORIDE ION, CHROMOSOMAL REPLICATION INITIATOR PROTEIN DNAA, ... | | Authors: | Natrajan, G, Noirot-Gros, M.-F, Zawilak-Pawlik, A, Kapp, U, Terradot, L. | | Deposit date: | 2009-07-31 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | The Structure of a Dnaa/Hoba Complex from Helicobacter Pylori Provides Insight Into Regulation of DNA Replication in Bacteria.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
