7KZQ
 
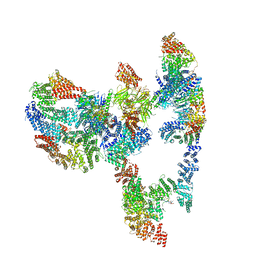 | | Structure of the human Fanconi anaemia Core-ID complex | | Descriptor: | E3 ubiquitin-protein ligase FANCL, Fanconi anemia core complex-associated protein 100, Fanconi anemia core complex-associated protein 20, ... | | Authors: | Wang, S.L, Pavletich, N.P. | | Deposit date: | 2020-12-10 | | Release date: | 2021-03-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of the FA core ubiquitin ligase closing the ID clamp on DNA.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KZS
 
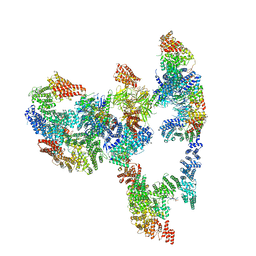 | |
1T6L
 
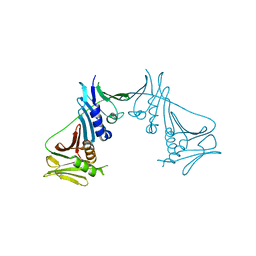 | | Crystal Structure of the Human Cytomegalovirus DNA Polymerase Subunit, UL44 | | Descriptor: | DNA polymerase processivity factor | | Authors: | Appleton, B.A, Loregian, A, Filman, D.J, Coen, D.M, Hogle, J.M. | | Deposit date: | 2004-05-06 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Cytomegalovirus DNA Polymerase Subunit UL44 Forms a C Clamp-Shaped Dimer.
Mol.Cell, 15, 2004
|
|
6GIS
 
 | | Structural basis of human clamp sliding on DNA | | Descriptor: | DNA (5'-D(P*AP*TP*AP*CP*GP*AP*TP*GP*GP*G)-3'), DNA (5'-D(P*CP*CP*CP*AP*TP*CP*GP*TP*AP*T)-3'), Proliferating cell nuclear antigen | | Authors: | De March, M, Merino, N, Barrera-Vilarmau, S, Crehuet, R, Onesti, S, Blanco, F.J, De Biasio, A. | | Deposit date: | 2018-05-15 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural basis of human PCNA sliding on DNA.
Nat Commun, 8, 2017
|
|
8DR0
 
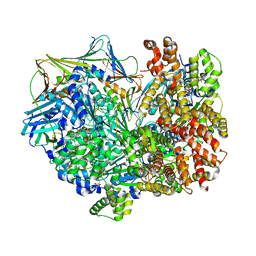 | | Closed state of RFC:PCNA bound to a 3' ss/dsDNA junction | | Descriptor: | DNA (5'-D(P*CP*CP*CP*CP*GP*GP*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*CP*GP*GP*GP*GP*GP*GP*GP*CP*CP*CP*CP*GP*GP*GP*G)-3'), GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
8DQZ
 
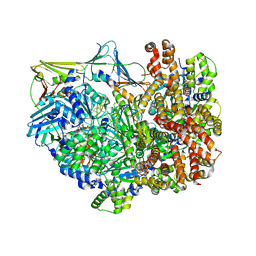 | | Intermediate state of RFC:PCNA bound to a 3' ss/dsDNA junction | | Descriptor: | DNA (5'-D(P*CP*CP*CP*CP*GP*GP*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*CP*GP*GP*GP*GP*GP*GP*GP*CP*CP*CP*CP*GP*GP*GP*G)-3'), GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
8DQX
 
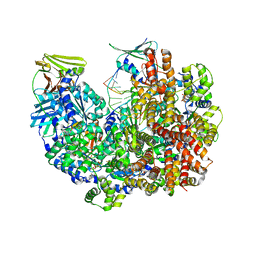 | | Open state of RFC:PCNA bound to a 3' ss/dsDNA junction | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*TP*T)-3'), DNA (5'-D(P*TP*CP*CP*GP*AP*GP*CP*GP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*GP*CP*CP*CP*GP*GP*A)-3'), ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
8DR6
 
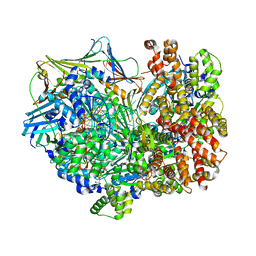 | | Closed state of RFC:PCNA bound to a nicked dsDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (32-MER), DNA (5'-D(P*CP*CP*CP*CP*CP*CP*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
8DR4
 
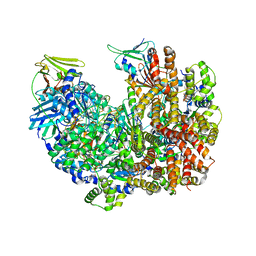 | | Open state of RFC:PCNA bound to a 3' ss/dsDNA junction (DNA2) without NTD | | Descriptor: | DNA (5'-D(P*AP*AP*GP*GP*GP*GP*GP*GP*GP*GP*GP*G)-3'), DNA (5'-D(P*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*TP*TP*T)-3'), DNA (5'-D(P*CP*CP*CP*CP*CP*CP*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
8DR7
 
 | | Open state of RFC:PCNA bound to a nicked dsDNA | | Descriptor: | DNA (26-MER), DNA (5'-D(P*AP*GP*GP*GP*GP*GP*GP*GP*GP*GP*G)-3'), DNA (5'-D(P*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
8DR3
 
 | | Closed state of RFC:PCNA bound to a 3' ss/dsDNA junction (DNA2) with NTD | | Descriptor: | DNA (5'-D(P*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*TP*TP*T)-3'), DNA (5'-D(P*CP*CP*CP*CP*CP*CP*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), DNA (5'-D(P*TP*TP*AP*GP*GP*GP*GP*GP*GP*GP*GP*GP*A)-3'), ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
6J8Y
 
 | | Crystal structure of the human RAD9-HUS1-RAD1-RHINO complex | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1, Checkpoint protein HUS1, ... | | Authors: | Hara, K, Iida, N, Sakurai, H, Hashimoto, H. | | Deposit date: | 2019-01-21 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the RAD9-RAD1-HUS1 checkpoint clamp bound to RHINO sheds light on the other side of the DNA clamp.
J.Biol.Chem., 295, 2020
|
|
2CHG
 
 | | Replication Factor C domains 1 and 2 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, REPLICATION FACTOR C SMALL SUBUNIT | | Authors: | Seybert, A, Singleton, M.R, Cook, N, Hall, D.R, Wigley, D.B. | | Deposit date: | 2006-03-14 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Communication between Subunits within an Archaeal Clamp-Loader Complex.
Embo J., 25, 2006
|
|
2AVT
 
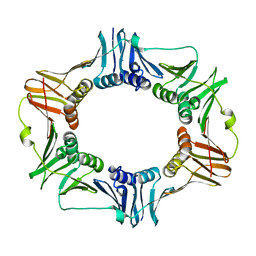 | | Crystal structure of the beta subunit from DNA polymerase of Streptococcus pyogenes | | Descriptor: | DNA polymerase III beta subunit | | Authors: | Argiriadi, M.A, Goedken, E.R, Bruck, I, O'donnell, M, Kuriyan, J. | | Deposit date: | 2005-08-30 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a DNA polymerase sliding clamp from a Gram-positive bacterium.
Bmc Struct.Biol., 6, 2006
|
|
5CXU
 
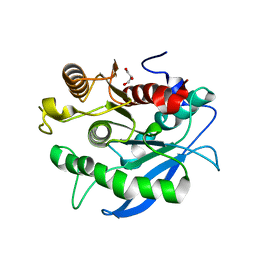 | |
5CXX
 
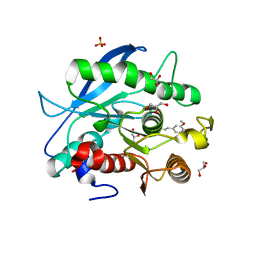 | | Structure of a CE1 ferulic acid esterase, AmCE1/Fae1A, from Anaeromyces mucronatus in complex with Ferulic acid | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Gruninger, R.J, Abbott, D.W. | | Deposit date: | 2015-07-29 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Contributions of a unique beta-clamp to substrate recognition illuminates the molecular basis of exolysis in ferulic acid esterases.
Biochem.J., 473, 2016
|
|
7STB
 
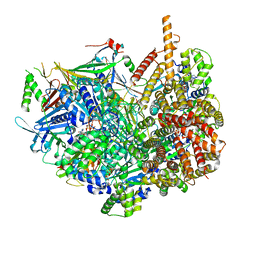 | | Closed state of Rad24-RFC:9-1-1 bound to a 5' ss/dsDNA junction | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DNA (5'-D(P*CP*GP*CP*TP*CP*CP*TP*TP*CP*CP*TP*GP*AP*CP*TP*CP*GP*TP*CP*C)-3'), ... | | Authors: | Castaneda, J.C, Schrecker, M, Remus, D, Hite, R.K. | | Deposit date: | 2021-11-12 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Mechanisms of loading and release of the 9-1-1 checkpoint clamp.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ST9
 
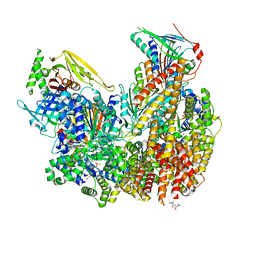 | | Open state of Rad24-RFC:9-1-1 bound to a 5' ss/dsDNA junction | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DNA (5'-D(P*CP*GP*CP*TP*CP*CP*TP*TP*CP*CP*TP*GP*AP*CP*TP*CP*GP*TP*CP*C)-3'), ... | | Authors: | Castaneda, J.C, Schrecker, M, Remus, D, Hite, R.K. | | Deposit date: | 2021-11-12 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Mechanisms of loading and release of the 9-1-1 checkpoint clamp.
Nat.Struct.Mol.Biol., 29, 2022
|
|
3G65
 
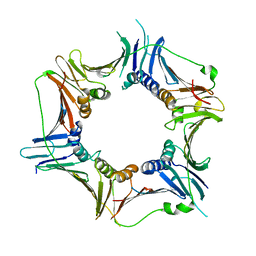 | | Crystal Structure of the Human Rad9-Rad1-Hus1 DNA Damage Checkpoint Complex | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1, Checkpoint protein HUS1 | | Authors: | Dore, A.S, Kilkenny, M.L, Rzechorzek, N.J, Pearl, L.H. | | Deposit date: | 2009-02-06 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the rad9-rad1-hus1 DNA damage checkpoint complex--implications for clamp loading and regulation.
Mol.Cell, 34, 2009
|
|
5I3H
 
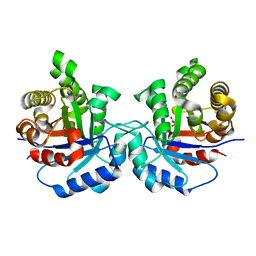 | | Structure-Function Studies on Role of Hydrophobic Clamping of a Basic Glutamate in Catalysis by Triosephosphate Isomerase | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, POTASSIUM ION, Triosephosphate isomerase, ... | | Authors: | Drake, E.J, Gulick, A.M, Richard, J.P, Zhai, X, Kim, K, Reinhardt, C.J. | | Deposit date: | 2016-02-10 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Function Studies of Hydrophobic Residues That Clamp a Basic Glutamate Side Chain during Catalysis by Triosephosphate Isomerase.
Biochemistry, 55, 2016
|
|
5I3F
 
 | | Structure-Function Studies on Role of Hydrophobic Clamping of a Basic Glutamate in Catalysis by Triosephosphate Isomerase | | Descriptor: | Triosephosphate isomerase, glycosomal | | Authors: | Drake, E.J, Gulick, A.M, Richard, J.P, Zhai, X, Kim, K, Reinhardt, C.J. | | Deposit date: | 2016-02-10 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure-Function Studies of Hydrophobic Residues That Clamp a Basic Glutamate Side Chain during Catalysis by Triosephosphate Isomerase.
Biochemistry, 55, 2016
|
|
5I3J
 
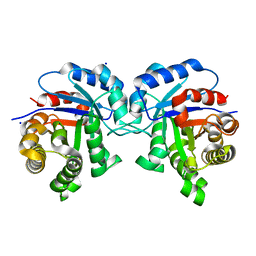 | | Structure-Function Studies on Role of Hydrophobic Clamping of a Basic Glutamate in Catalysis by Triosephosphate Isomerase | | Descriptor: | SODIUM ION, Triosephosphate isomerase, glycosomal | | Authors: | Drake, E.J, Gulick, A.M, Richard, J.P, Zhai, X, Kim, K, Reinhardt, C.J. | | Deposit date: | 2016-02-10 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Function Studies of Hydrophobic Residues That Clamp a Basic Glutamate Side Chain during Catalysis by Triosephosphate Isomerase.
Biochemistry, 55, 2016
|
|
5I3K
 
 | | Structure-Function Studies on Role of Hydrophobic Clamping of a Basic Glutamate in Catalysis by Triosephosphate Isomerase | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, SODIUM ION, Triosephosphate isomerase, ... | | Authors: | Drake, E.J, Gulick, A.M, Richard, J.P, Zhai, X, Kim, K, Reinhardt, C.J. | | Deposit date: | 2016-02-10 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.209 Å) | | Cite: | Structure-Function Studies of Hydrophobic Residues That Clamp a Basic Glutamate Side Chain during Catalysis by Triosephosphate Isomerase.
Biochemistry, 55, 2016
|
|
5I3G
 
 | | Structure-Function Studies on Role of Hydrophobic Clamping of a Basic Glutamate in Catalysis by Triosephosphate Isomerase | | Descriptor: | Triosephosphate isomerase, glycosomal | | Authors: | Drake, E.J, Gulick, A.M, Richard, J.P, Zhai, X, Kim, K, Reinhardt, C.J. | | Deposit date: | 2016-02-10 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-Function Studies of Hydrophobic Residues That Clamp a Basic Glutamate Side Chain during Catalysis by Triosephosphate Isomerase.
Biochemistry, 55, 2016
|
|
5I3I
 
 | | Structure-Function Studies on Role of Hydrophobic Clamping of a Basic Glutamate in Catalysis by Triosephosphate Isomerase | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate isomerase, glycosomal | | Authors: | Drake, E.J, Gulick, A.M, Richard, J.P, Zhai, X, Kim, K, Reinhardt, C.J. | | Deposit date: | 2016-02-10 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Function Studies of Hydrophobic Residues That Clamp a Basic Glutamate Side Chain during Catalysis by Triosephosphate Isomerase.
Biochemistry, 55, 2016
|
|
