1NXB
 
 | |
1YVT
 
 | | The high salt (phosphate) crystal structure of CO Hemoglobin E (Glu26Lys) at physiological pH (pH 7.35) | | Descriptor: | CARBON MONOXIDE, GLYCEROL, Hemoglobin alpha chain, ... | | Authors: | Malashkevich, V.N, Balazs, T.C, Almo, S.C, Hirsch, R.E. | | Deposit date: | 2005-02-16 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The high salt (phosphate) crystal structure of CO Hemoglobin E (Glu26Lys) at physiological pH (pH 7.35)
To be Published
|
|
1JY7
 
 | | THE STRUCTURE OF HUMAN METHEMOGLOBIN. THE VARIATION OF A THEME | | Descriptor: | Hemoglobin alpha chain, Hemoglobin beta chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Biswal, B.K, Vijayan, M. | | Deposit date: | 2001-09-11 | | Release date: | 2002-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of human oxy- and deoxyhaemoglobin at different levels of humidity: variability in the T state.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1JZ7
 
 | |
1ZM6
 
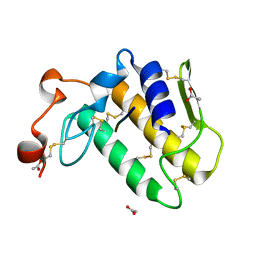 | | Crystal structure of the complex formed beween a group I phospholipase A2 and designed penta peptide Leu-Ala-Ile-Tyr-Ser at 2.6A resolution | | Descriptor: | ACETATE ION, Phospholipase A2 isoform 3, designed penta peptide Leu-Ala-Ile-Tyr-Ser | | Authors: | Singh, R.K, Singh, N, Jabeen, T, Sharma, S, Dey, S, Singh, T.P. | | Deposit date: | 2005-05-10 | | Release date: | 2005-06-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the complex of group I PLA2 with a group II-specific peptide Leu-Ala-Ile-Tyr-Ser (LAIYS) at 2.6 A resolution.
J.Drug Target., 13, 2005
|
|
1JZ4
 
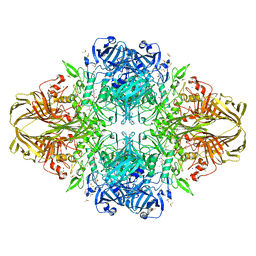 | |
1K2R
 
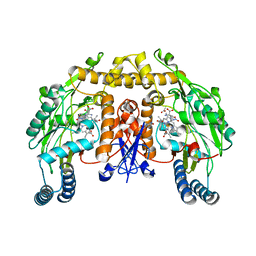 | | Structure of rat brain nNOS heme domain complexed with NG-nitro-L-arginine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-OMEGA-NITRO-L-ARGININE, ... | | Authors: | Li, H, Martasek, P, Masters, B.S.S, Poulos, T.L, Raman, C.S. | | Deposit date: | 2001-09-28 | | Release date: | 2003-03-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of rat brain nNOS heme domain
To be Published
|
|
1JPS
 
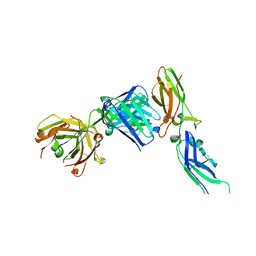 | | Crystal structure of tissue factor in complex with humanized Fab D3h44 | | Descriptor: | immunoglobulin Fab D3H44, heavy chain, light chain, ... | | Authors: | Faelber, K, Kirchhofer, D, Presta, L, Kelley, R.F, Muller, Y.A. | | Deposit date: | 2001-08-03 | | Release date: | 2002-02-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The 1.85 A resolution crystal structures of tissue factor in complex with humanized Fab D3h44 and of free humanized Fab D3h44: revisiting the solvation of antigen combining sites.
J.Mol.Biol., 313, 2001
|
|
1Z3S
 
 | |
1OPB
 
 | |
1OQM
 
 | | A 1:1 complex between alpha-lactalbumin and beta1,4-galactosyltransferase in the presence of UDP-N-acetyl-galactosamine | | Descriptor: | Alpha-lactalbumin, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Ramakrishnan, B, Qasba, P.K. | | Deposit date: | 2003-03-10 | | Release date: | 2003-03-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based design of beta 1,4-galactosyltransferase I (beta 4Gal-T1) with equally efficient
N-acetylgalactosaminyltransferase activity: point mutation broadens beta 4Gal-T1 donor specificity.
J.Biol.Chem., 277, 2002
|
|
1JTA
 
 | | Crystal Structure of Pectate Lyase A (C2 form) | | Descriptor: | SULFATE ION, pectate lyase A | | Authors: | Thomas, L.M, Doan, C.N, Oliver, R.L, Yoder, M.D. | | Deposit date: | 2001-08-20 | | Release date: | 2002-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of pectate lyase A: comparison to other isoforms.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1OUQ
 
 | | Crystal structure of wild-type Cre recombinase-loxP synapse | | Descriptor: | Cre recombinase, IODIDE ION, MAGNESIUM ION, ... | | Authors: | Ennifar, E, Meyer, J.E.W, Buchholz, F, Stewart, A.F, Suck, D. | | Deposit date: | 2003-03-25 | | Release date: | 2003-09-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a wild-type Cre recombinase-loxP synapse reveals a novel spacer conformation suggesting an alternative mechanism for DNA cleavage activation
Nucleic Acids Res., 31, 2003
|
|
1JIB
 
 | | Complex of Alpha-amylase II (TVA II) from Thermoactinomyces vulgaris R-47 with Maltotetraose Based on a Crystal Soaked with Maltohexaose. | | Descriptor: | NEOPULLULANASE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yokota, T, Tonozuka, T, Shimura, Y, Ichikawa, K, Kamitori, S, Sakano, Y. | | Deposit date: | 2001-07-02 | | Release date: | 2001-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of Thermoactinomyces vulgaris R-47 alpha-amylase II complexed with substrate analogues.
Biosci.Biotechnol.Biochem., 65, 2001
|
|
1OTS
 
 | | Structure of the Escherichia coli ClC Chloride channel and Fab Complex | | Descriptor: | CHLORIDE ION, Fab fragment (heavy chain), Fab fragment (light chain), ... | | Authors: | Dutzler, R, Campbell, E.B, MacKinnon, R. | | Deposit date: | 2003-03-22 | | Release date: | 2003-04-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Gating the Selectivity Filter in ClC Chloride Channels
Science, 300, 2003
|
|
1Z99
 
 | | Solution structure of Crotamine, a myotoxin from Crotalus durissus terrificus | | Descriptor: | Crotamine | | Authors: | Fadel, V, Bettendorff, P, Herrmann, T, de Azevedo, W.F, Oliveira, E.B, Yamane, T, Wuthrich, K. | | Deposit date: | 2005-04-01 | | Release date: | 2006-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Automated NMR structure determination and disulfide bond identification of the myotoxin crotamine from Crotalus durissus terrificus.
Toxicon, 46, 2005
|
|
1OWN
 
 | | DATA3:DNA photolyase / received X-rays dose 4.8 exp15 photons/mm2 | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1JLC
 
 | | CRYSTAL STRUCTURE OF Y181C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH PETT-2 | | Descriptor: | HIV-1 RT A-chain, HIV-1 RT B-chain, N-[[3-FLUORO-4-ETHOXY-PYRID-2-YL]ETHYL]-N'-[5-CHLORO-PYRIDYL]-THIOUREA | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
1Z3Z
 
 | | The crystal structure of a DGD mutant: Q52A | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Fogle, E.J, Liu, W, Toney, M.D. | | Deposit date: | 2005-03-14 | | Release date: | 2006-01-03 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Role of q52 in catalysis of decarboxylation and transamination in dialkylglycine decarboxylase.
Biochemistry, 44, 2005
|
|
1JLU
 
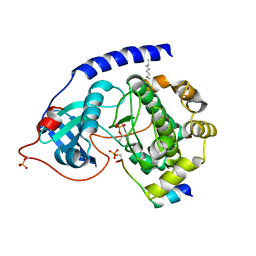 | | Crystal Structure of the Catalytic Subunit of cAMP-dependent Protein Kinase Complexed with a Phosphorylated Substrate Peptide and Detergent | | Descriptor: | AMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR, ... | | Authors: | Madhusudan, Trafny, E.A, Xuong, N.-H, Adams, J.A, Ten Eyck, L.F, Taylor, S.S, Sowadski, J.M. | | Deposit date: | 2001-07-16 | | Release date: | 2001-08-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | cAMP-dependent protein kinase: crystallographic insights into substrate recognition and phosphotransfer.
Protein Sci., 3, 1994
|
|
1Z4L
 
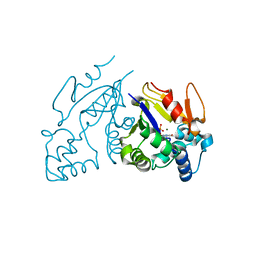 | | Structure of the D41N variant of the human mitochondrial deoxyribonucleotidase in complex with thymidine 5'-monophosphate | | Descriptor: | 5'(3')-deoxyribonucleotidase, MAGNESIUM ION, THYMIDINE-5'-PHOSPHATE | | Authors: | Wallden, K, Ruzzenente, B, Rinaldo-Matthis, A, Bianchi, V, Nordlund, P. | | Deposit date: | 2005-03-16 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for substrate specificity of the human mitochondrial deoxyribonucleotidase
STRUCTURE, 13, 2005
|
|
1Z5H
 
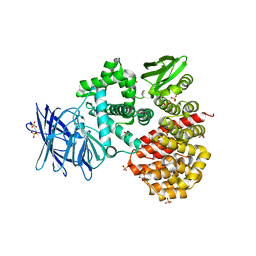 | | Crystal structures of the Tricorn interacting Factor F3 from Thermoplasma acidophilum | | Descriptor: | SULFATE ION, Tricorn protease interacting factor F3, ZINC ION | | Authors: | Kyrieleis, O.J.P, Goettig, P, Kiefersauer, R, Huber, R, Brandstetter, H. | | Deposit date: | 2005-03-18 | | Release date: | 2005-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the Tricorn Interacting Factor F3 from Thermoplasma acidophilum, a Zinc Aminopeptidase in Three Different Conformations
J.MOL.BIOL., 349, 2005
|
|
1JO5
 
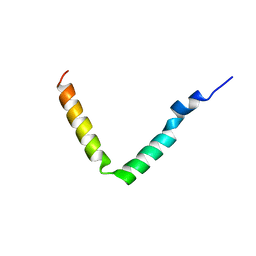 | | Rhodobacter sphaeroides Light Harvesting 1 beta Subunit in Detergent Micelles | | Descriptor: | LIGHT-HARVESTING PROTEIN B-875 | | Authors: | Sorgen, P.L, Cahill, S.M, Krueger-Koplin, R.D, Krueger-Koplin, S.T, Schenck, C.G, Girvin, M.E. | | Deposit date: | 2001-07-26 | | Release date: | 2002-02-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Rhodobacter sphaeroides light-harvesting 1 beta subunit in detergent micelles.
Biochemistry, 41, 2002
|
|
1Z6O
 
 | | Crystal Structure of Trichoplusia ni secreted ferritin | | Descriptor: | CALCIUM ION, FE (III) ION, Ferritin heavy chain, ... | | Authors: | Hamburger, A.E, West Jr, A.P, Hamburger, Z.A, Hamburger, P, Bjorkman, P.J. | | Deposit date: | 2005-03-22 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of a secreted insect ferritin reveals a symmetrical arrangement of heavy and light chains.
J.Mol.Biol., 349, 2005
|
|
1ZAH
 
 | | Fructose-1,6-bisphosphate aldolase from rabbit muscle | | Descriptor: | Fructose-bisphosphate aldolase A | | Authors: | St-Jean, M, Lafrance-Vanasse, J, Liotard, B, Sygusch, J. | | Deposit date: | 2005-04-06 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High Resolution Reaction Intermediates of Rabbit Muscle Fructose-1,6-bisphosphate Aldolase: substrate cleavage and induced fit.
J.Biol.Chem., 280, 2005
|
|
