3V5D
 
 | | HLA-A2.1 KVAELVHFL | | Descriptor: | Beta-2-microglobulin, HIV peptide KVAELVHFL, HLA class I histocompatibility antigen, ... | | Authors: | Collins, E.J, Lee, H.Y. | | Deposit date: | 2011-12-16 | | Release date: | 2012-12-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prediction of conformation and immunogenicity of peptides bound to MHC molecules
To be Published
|
|
6HOJ
 
 | | Structure of Beclin1 LIR motif bound to GABARAP | | Descriptor: | 1,2-ETHANEDIOL, Beclin-1,Gamma-aminobutyric acid receptor-associated protein, SULFATE ION | | Authors: | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
6HPF
 
 | | Structure of Inactive E165Q mutant of fungal non-CBM carrying GH26 endo-b-mannanase from Yunnania penicillata in complex with alpha-62-61-di-galactosyl-mannotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, CHLORIDE ION, ... | | Authors: | von Freiesleben, P, Moroz, O.V, Blagova, E, Wiemann, M, Spodsberg, N, Agger, J.W, Davies, G.J, Wilson, K.S, Stalbrand, H, Meyer, A.S, Krogh, K.B.R.M. | | Deposit date: | 2018-09-20 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal structure and substrate interactions of an unusual fungal non-CBM carrying GH26 endo-beta-mannanase from Yunnania penicillata.
Sci Rep, 9, 2019
|
|
4ZNS
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in complex with a 3-Fluoro-substituted OBHS derivative | | Descriptor: | 3-fluorophenyl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate, Estrogen receptor, Nuclear receptor-interacting peptide | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-05-05 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
3V5K
 
 | | HLA2.1 KVAELVWFL | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HIV-based altered-peptide ligand KVAELVWFL, ... | | Authors: | Collins, E.J, Lee, H.Y. | | Deposit date: | 2011-12-16 | | Release date: | 2012-12-19 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Prediction of Immunogenicity of altered-peptide ligands to HIV bound to MHC
To be Published
|
|
1HWV
 
 | | MOLECULAR TOPOLOGY OF POLYCYCLIC AROMATIC CARCINOGENS DETERMINES DNA ADDUCT CONFORMATION: A LINK TO TUMORIGENIC ACTIVITY | | Descriptor: | (1S)-1,2,3,4-TETRAHYDRO-BENZO[C]PHENANTHRENE-2,3,4-TRIOL, 5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3' | | Authors: | Patel, D.J, Lin, C.H, Geacintov, N.E, Broyde, S, Huang, X, Kolbanovskii, A, Hingerty, B.E, Amin, S. | | Deposit date: | 2001-01-10 | | Release date: | 2001-03-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular topology of polycyclic aromatic carcinogens determines DNA adduct conformation: a link to tumorigenic activity.
J.Mol.Biol., 306, 2001
|
|
1HX4
 
 | | MOLECULAR TOPOLOGY OF POLYCYCLIC AROMATIC CARCINOGENS DETERMINES DNA ADDUCT CONFORMATION: A LINK TO TUMORIGENIC ACTIVITY | | Descriptor: | (1R)-1,2,3,4-TETRAHYDRO-BENZO[C]PHENANTHRENE-2,3,4-TRIOL, 5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3' | | Authors: | Patel, D.J, Lin, C.H, Geacintov, N.E, Broyde, S, Huang, X, Kolbanovskii, A, Hingerty, B.E, Amin, S. | | Deposit date: | 2001-01-11 | | Release date: | 2001-03-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular topology of polycyclic aromatic carcinogens determines DNA adduct conformation: a link to tumorigenic activity.
J.Mol.Biol., 306, 2001
|
|
6HT1
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with SGC-iMLLT (compound 92) | | Descriptor: | 1,2-ETHANEDIOL, 1-methyl-~{N}-[2-[[(2~{S})-2-methylpyrrolidin-1-yl]methyl]-3~{H}-benzimidazol-5-yl]indazole-5-carboxamide, Protein ENL, ... | | Authors: | Heidenreich, D, Chaikuad, A, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-10-02 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of an MLLT1/3 YEATS Domain Chemical Probe.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
2G6X
 
 | |
5OWX
 
 | | Inside-out FMDV A10 capsid | | Descriptor: | Genome polyprotein | | Authors: | Kotecha, A, Malik, N, Fry, E.E, Stuart, D.I. | | Deposit date: | 2017-09-04 | | Release date: | 2017-09-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Structures of foot and mouth disease virus pentamers: Insight into capsid dissociation and unexpected pentamer reassociation.
PLoS Pathog., 13, 2017
|
|
8EHK
 
 | |
4K80
 
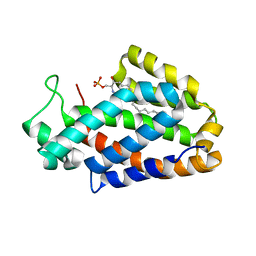 | | Crystal structure of human ceramide-1-phosphate transfer protein (CPTP) in complex with 2:0 ceramide-1-phosphate (2:0-C1P) | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Glycolipid transfer protein domain-containing protein 1 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-04-17 | | Release date: | 2013-07-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids.
Nature, 500, 2013
|
|
8EHM
 
 | |
8EHL
 
 | |
8EHJ
 
 | |
6D8E
 
 | |
7NCB
 
 | | Glutathione-S-transferase GliG mutant H26A | | Descriptor: | Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NC2
 
 | |
1CE9
 
 | | HELIX CAPPING IN THE GCN4 LEUCINE ZIPPER | | Descriptor: | PROTEIN (GCN4-PMSE) | | Authors: | Lu, M, Shu, W, Ji, H, Spek, E, Wang, L.-Y, Kallenbach, N.R. | | Deposit date: | 1999-03-18 | | Release date: | 1999-03-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Helix capping in the GCN4 leucine zipper.
J.Mol.Biol., 288, 1999
|
|
2QX0
 
 | | Crystal Structure of Yersinia pestis HPPK (Ternary Complex) | | Descriptor: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, 7,8-dihydro-6-hydroxymethylpterin-pyrophosphokinase, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Blaszczyk, J, Cherry, S, Tropea, J.E, Waugh, D.S, Ji, X. | | Deposit date: | 2007-08-10 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and activity of Yersinia pestis 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase as a novel target for the development of antiplague therapeutics.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2QEP
 
 | | Crystal structure of the D1 domain of PTPRN2 (IA2beta) | | Descriptor: | CHLORIDE ION, Receptor-type tyrosine-protein phosphatase N2 | | Authors: | Ugochukwu, E, Barr, A, Alfano, I, Berridge, G, Burgess-Brown, N, Das, S, Fedorov, O, King, O, Niesen, F, Watt, S, Savitsky, P, Salah, E, Pike, A.C.W, Bunkoczi, G, von Delft, F, Sundstrom, M, Edwards, A, Arrowsmith, C.H, Weigelt, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-26 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
4GAO
 
 | | DCNL complex with N-terminally acetylated NEDD8 E2 peptide | | Descriptor: | BROMIDE ION, DCN1-like protein 2, NEDD8-conjugating enzyme Ubc12 | | Authors: | Monda, J.K, Scott, D.C, Miller, D.J, Harper, J.W, Bennett, E.J, Schulman, B.A. | | Deposit date: | 2012-07-25 | | Release date: | 2012-11-28 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Structural Conservation of Distinctive N-terminal Acetylation-Dependent Interactions across a Family of Mammalian NEDD8 Ligation Enzymes.
Structure, 21, 2013
|
|
3LIK
 
 | | Human MMP12 in complex with non-zinc chelating inhibitor | | Descriptor: | ACETOHYDROXAMIC ACID, CALCIUM ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Stura, E.A, Dive, V, Devel, L, Czarny, B, Vera, L, Beau, F. | | Deposit date: | 2010-01-25 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights from selective non-phosphinic inhibitors of MMP-12 tailored to fit with an S1' loop canonical conformation.
J.Biol.Chem., 285, 2010
|
|
6DDW
 
 | | Mycobacterium tuberculosis Dihydrofolate Reductase complexed with beta-NADPH and N-(4-{[(2-amino-4-oxo-3,4-dihydropteridin-6-yl)methyl]amino}-2-hydroxybenzene-1-carbonyl)-L-glutamic acid | | Descriptor: | Dihydrofolate reductase, GLYCEROL, N-(4-{[(2-amino-4-oxo-3,4-dihydropteridin-6-yl)methyl]amino}-2-hydroxybenzene-1-carbonyl)-L-glutamic acid, ... | | Authors: | Hajian, B, Scocchera, E, Wright, D. | | Deposit date: | 2018-05-10 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Drugging the Folate Pathway in Mycobacterium tuberculosis: The Role of Multi-targeting Agents.
Cell Chem Biol, 26, 2019
|
|
7NPT
 
 | | Cytosolic bridge of an intact ESX-5 inner membrane complex | | Descriptor: | ESX-5 secretion system protein EccC5, ESX-5 secretion system protein EccD5 | | Authors: | Fahrenkamp, D, Bunduc, C.M, Wald, J, Ummels, R, Bitter, W, Houben, E.N.G, Marlovits, T.C. | | Deposit date: | 2021-02-28 | | Release date: | 2021-05-26 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structure and dynamics of a mycobacterial type VII secretion system.
Nature, 593, 2021
|
|
