6N2G
 
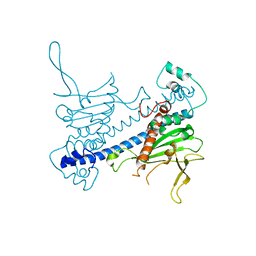 | | Crystal structure of Caenorhabditis elegans NAP1 | | Descriptor: | Nucleosome Assembly Protein | | Authors: | Bhattacharyya, S, DArcy, S. | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Characterization of Caenorhabditis elegans Nucleosome Assembly Protein 1 Uncovers the Role of Acidic Tails in Histone Binding.
Biochemistry, 58, 2019
|
|
6MNJ
 
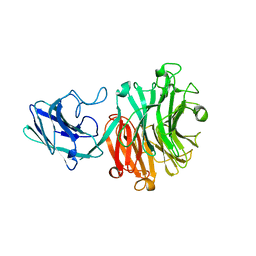 | | Hadza microbial sialidase Hz136 | | Descriptor: | Hz136 | | Authors: | Rees, S.D, Zaramela, L, Zengler, K, Chang, G. | | Deposit date: | 2018-10-01 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Gut bacteria responding to dietary change encode sialidases that exhibit preference for red meat-associated carbohydrates.
Nat Microbiol, 4, 2019
|
|
6MRV
 
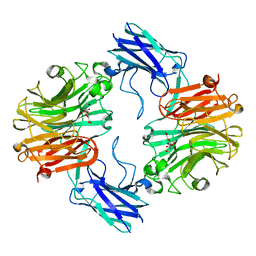 | | Sialidase26 co-crystallized with DANA | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, Sialidase26 | | Authors: | Zaramela, L.S, Martino, C, Alisson-Silva, F, Rees, S.D, Diaz, S.L, Chuzel, L, Ganatra, M.B, Taron, C.H, Zuniga, C, Chang, G, Varki, A, Zengler, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Gut bacteria responding to dietary change encode sialidases that exhibit preference for red meat-associated carbohydrates.
Nat Microbiol, 4, 2019
|
|
6MPH
 
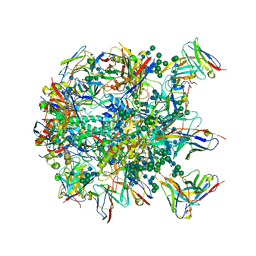 | | Cryo-EM structure at 3.8 A resolution of HIV-1 fusion peptide-directed antibody, DF1W-a.01, elicited by vaccination of Rhesus macaques, in complex with stabilized HIV-1 Env BG505 DS-SOSIP, which was also bound to antibodies VRC03 and PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DF1W-a.01 Light chain, ... | | Authors: | Acharya, P, Xu, K, Kwong, P.D. | | Deposit date: | 2018-10-06 | | Release date: | 2019-07-24 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Antibody Lineages with Vaccine-Induced Antigen-Binding Hotspots Develop Broad HIV Neutralization.
Cell, 178, 2019
|
|
4ZRR
 
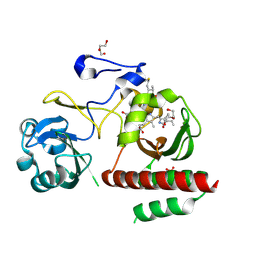 | | Crystal Structure of Monomeric Bacteriophytochrome mutant D207L Y263F at 1.5 A resolution Using a home source. | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, 3-[2-[(Z)-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-1-ium-2-ylidene]methyl]-5-[(Z)-[(3E,4R)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Bacteriophytochrome, ... | | Authors: | Bhattacharya, S, Satyshur, K.A, Lehtivuori, H, Forest, K.T. | | Deposit date: | 2015-05-12 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Removal of Chromophore-Proximal Polar Atoms Decreases Water Content and Increases Fluorescence in a Near Infrared Phytofluor.
Front Mol Biosci, 2, 2015
|
|
6MPG
 
 | | Cryo-EM structure at 3.2 A resolution of HIV-1 fusion peptide-directed antibody, A12V163-b.01, elicited by vaccination of Rhesus macaques, in complex with stabilized HIV-1 Env BG505 DS-SOSIP, which was also bound to antibodies VRC03 and PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, A12V163-b.01 Heavy Chain, ... | | Authors: | Acharya, P, Kwong, P.D. | | Deposit date: | 2018-10-06 | | Release date: | 2019-07-24 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Antibody Lineages with Vaccine-Induced Antigen-Binding Hotspots Develop Broad HIV Neutralization.
Cell, 178, 2019
|
|
6MRX
 
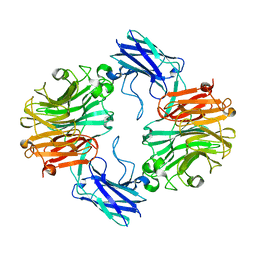 | | Sialidase26 apo | | Descriptor: | Sialidase26 | | Authors: | Zaramela, L.S, Martino, C, Alisson-Silva, F, Rees, S.D, Diaz, S.L, Chuzel, L, Ganatra, M.B, Taron, C.H, Zuniga, C, Chang, G, Varki, A, Zengler, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Gut bacteria responding to dietary change encode sialidases that exhibit preference for red meat-associated carbohydrates.
Nat Microbiol, 4, 2019
|
|
6MYV
 
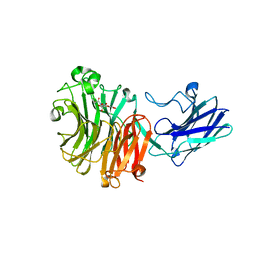 | | Sialidase26 co-crystallized with DANA-Gc | | Descriptor: | 2,6-anhydro-3,5-dideoxy-5-[(hydroxyacetyl)amino]-D-glycero-L-altro-non-2-enonic acid, Sialidase26 | | Authors: | Zaramela, L.S, Martino, C, Alisson-Silva, F, Rees, S.D, Diaz, S.L, Chuzel, L, Ganatra, M.B, Taron, C.H, Zuniga, C, Huang, J, Siegel, D, Chang, G, Varki, A, Zengler, K. | | Deposit date: | 2018-11-02 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Gut bacteria responding to dietary change encode sialidases that exhibit preference for red meat-associated carbohydrates.
Nat Microbiol, 4, 2019
|
|
1ONY
 
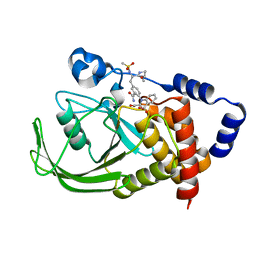 | | Oxalyl-Aryl-Amino Benzoic Acid inhibitors of PTP1B, compound 17 | | Descriptor: | 2-{[2-(2-CARBAMOYL-VINYL)-4-(2-METHANESULFONYLAMINO-2-PENTYLCARBAMOYL-ETHYL)-PHENYL]-OXALYL-AMINO}-BENZOIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Liu, G, Szczepankiewicz, B.G, Pei, Z, Janowich, D.A, Xin, Z, Hadjuk, P.J, Abad-Zapatero, C, Liang, H, Hutchins, C.W, Fesik, S.W, Ballaron, S.J, Stashko, M.A, Lubben, T, Mika, A.K, Zinker, B.A, Trevillyan, J.M, Jirousek, M.R. | | Deposit date: | 2003-03-02 | | Release date: | 2003-05-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery and Structure-Activity Relationship of Oxalylarylaminobenzoic
Acids as Inhibitors of Protein Tyrosine Phosphatase 1B
J.Med.Chem., 46, 2003
|
|
1FYC
 
 | | INNER LIPOYL DOMAIN FROM HUMAN PYRUVATE DEHYDROGENASE (PDH) COMPLEX, NMR, 1 STRUCTURE | | Descriptor: | DIHYDROLIPOAMIDE ACETYLTRANSFERASE (E2P) | | Authors: | Howard, M.J, Fuller, C, Broadhurst, R.W, Quinn, J, Yeaman, S.J, Perham, R.N. | | Deposit date: | 1997-02-21 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the major autoantigen in primary biliary cirrhosis.
Gastroenterology, 115, 1998
|
|
4KA2
 
 | |
3AAG
 
 | | Crystal structure of C. jejuni pglb C-terminal domain | | Descriptor: | CALCIUM ION, General glycosylation pathway protein | | Authors: | Maita, N, Kohda, D. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Comparative structural biology of Eubacterial and Archaeal oligosaccharyltransferases.
J.Biol.Chem., 285, 2010
|
|
6QSP
 
 | | Ketosynthase (ApeO) in Complex with its Chain Length Factor (ApeC) from Xenorhabdus doucetiae | | Descriptor: | Beta-ketoacyl synthase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Grammbitter, G.L.C, Schmalhofer, M, Groll, M, Bode, H. | | Deposit date: | 2019-02-21 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | An Uncommon Type II PKS Catalyzes Biosynthesis of Aryl Polyene Pigments.
J.Am.Chem.Soc., 141, 2019
|
|
3BKQ
 
 | |
6QSR
 
 | | The Dehydratase Heterocomplex ApeI:P from Xenorhabdus doucetiae | | Descriptor: | AMP-dependent synthetase and ligase, Beta-hydroxyacyl-(Acyl-carrier-protein) dehydratase FabA/FabZ, SODIUM ION | | Authors: | Grammbitter, G.L.C, Schmalhofer, M, Groll, M, Bode, H. | | Deposit date: | 2019-02-21 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An Uncommon Type II PKS Catalyzes Biosynthesis of Aryl Polyene Pigments.
J.Am.Chem.Soc., 141, 2019
|
|
3C04
 
 | |
3V6I
 
 | | Crystal structure of the peripheral stalk of Thermus thermophilus H+-ATPase/synthase at 2.25 A resolution | | Descriptor: | CALCIUM ION, SODIUM ION, V-type ATP synthase subunit E, ... | | Authors: | Stewart, A.G, Lee, L.K, Donohoe, M, Chaston, J.J, Stock, D. | | Deposit date: | 2011-12-19 | | Release date: | 2012-02-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The dynamic stator stalk of rotary ATPases
Nat Commun, 3, 2012
|
|
4OL4
 
 | | Crystal structure of secreted proline rich antigen MTC28 (Rv0040c) from Mycobacterium tuberculosis | | Descriptor: | Proline-rich 28 kDa antigen | | Authors: | Kundu, P, Biswas, R, Mukherjee, S, Reinhard, L, Mueller-dieckmann, J, Weiss, M.S, Das, A.K. | | Deposit date: | 2014-01-23 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based Epitope Mapping of Mycobacterium tuberculosis Secretary Antigen MTC28
J.Biol.Chem., 291, 2016
|
|
1PW3
 
 | | Crystal structure of JtoR68S | | Descriptor: | CADMIUM ION, immunoglobulin lambda chain variable region | | Authors: | Dealwis, C, Gupta, V, Wilkerson, M. | | Deposit date: | 2003-06-30 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of light chain amyloidogenicity: comparison of the thermodynamic properties, fibrillogenic potential and tertiary structural features of four Vlambda6 proteins.
J.Mol.Recog., 17, 2004
|
|
4PWS
 
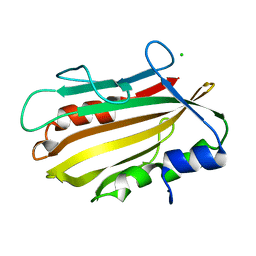 | | Crystal structure of secreted proline rich antigen MTC28 (Rv0040c) at 2.15 A with bound chloride from Mycobacterium tuberculosis | | Descriptor: | CHLORIDE ION, Proline-rich 28 kDa antigen | | Authors: | Kundu, P, Biswas, R, Mukherjee, S, Reinhard, L, Mueller-dieckmann, J, Weiss, M.S, Das, A.K. | | Deposit date: | 2014-03-21 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-based Epitope Mapping of Mycobacterium tuberculosis Secretary Antigen MTC28
J.Biol.Chem., 291, 2016
|
|
1PEW
 
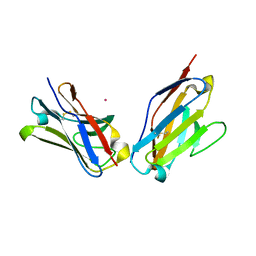 | | High Resolution Crystal Structure of Jto2, a mutant of the non-amyloidogenic Lamba6 Light Chain, Jto | | Descriptor: | CADMIUM ION, Jto2, a LAMBDA-6 TYPE IMMUNOGLOBULIN LIGHT CHAIN, ... | | Authors: | Dealwis, C, Gupta, V, Wilkerson, M. | | Deposit date: | 2003-05-22 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of light chain amyloidogenicity: comparison of the thermodynamic properties, fibrillogenic potential and tertiary structural features of four V(lambda)6 proteins
J.Mol.Recog., 17, 2004
|
|
5OLY
 
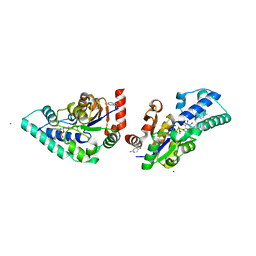 | |
1DV5
 
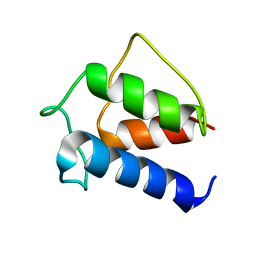 | | TERTIARY STRUCTURE OF APO-D-ALANYL CARRIER PROTEIN | | Descriptor: | APO-D-ALANYL CARRIER PROTEIN | | Authors: | Volkman, B.F, Zhang, Q, Debabov, D.V, Rivera, E, Kresheck, G.C, Neuhaus, F.C. | | Deposit date: | 2000-01-19 | | Release date: | 2001-08-01 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Biosynthesis of D-alanyl-lipoteichoic acid: the tertiary structure of apo-D-alanyl carrier protein.
Biochemistry, 40, 2001
|
|
5OLX
 
 | |
5OLW
 
 | |
