1SLO
 
 | | FIRST STEM LOOP OF THE SL1 RNA FROM CAENORHABDITIS ELEGANS, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | RNA (5'-R(*UP*UP*AP*CP*CP*CP*AP*AP*GP*UP*UP*UP*GP*AP*GP*GP*UP*AP*A)-3') | | Authors: | Greenbaum, N.L, Radhakrishnan, I, Patel, D.J, Hirsh, D. | | Deposit date: | 1996-05-24 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the donor site of a trans-splicing RNA.
Structure, 4, 1996
|
|
1SLP
 
 | | FIRST STEM LOOP OF THE SL1 RNA FROM CAENORHABDITIS ELEGANS, NMR, 16 STRUCTURES | | Descriptor: | RNA (5'-R(*UP*UP*AP*CP*CP*CP*AP*AP*GP*UP*UP*UP*GP*AP*GP*GP*UP*AP*A)-3') | | Authors: | Greenbaum, N.L, Radhakrishnan, I, Patel, D.J, Hirsh, D. | | Deposit date: | 1996-05-24 | | Release date: | 1997-04-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the donor site of a trans-splicing RNA.
Structure, 4, 1996
|
|
1N8X
 
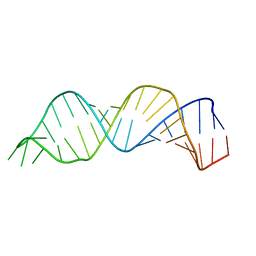 | | Solution structure of HIV-1 Stem Loop SL1 | | Descriptor: | HIV-1 STEM LOOP SL1 MONOMERIC RNA | | Authors: | Lawrence, D.C, Stover, C.C, Noznitsky, J, Wu, Z, Summers, M.F. | | Deposit date: | 2002-11-21 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Intact Stem and Bulge of HIV-1 Psi-RNA Stem-Loop SL1
J.Mol.Biol., 326, 2003
|
|
2JQ7
 
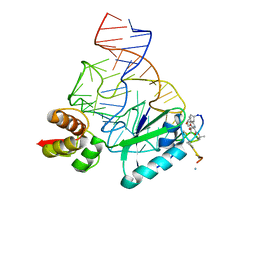 | | Model for thiostrepton binding to the ribosomal L11-RNA | | Descriptor: | 50S RIBOSOMAL PROTEIN L11, RIBOSOMAL RNA, THIOSTREPTON | | Authors: | Jonker, H.R.A, Ilin, S, Grimm, S.K, Woehnert, J, Schwalbe, H. | | Deposit date: | 2007-05-30 | | Release date: | 2007-07-03 | | Last modified: | 2021-08-18 | | Method: | SOLUTION NMR | | Cite: | L11 Domain Rearrangement Upon Binding to RNA and Thiostrepton Studied by NMR Spectroscopy
Nucleic Acids Res., 35, 2007
|
|
3WZI
 
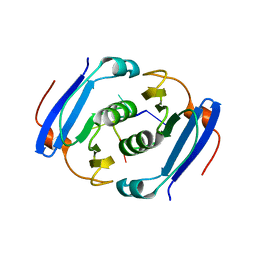 | | Crystal structure of AfCsx3 in complex with ssRNA | | Descriptor: | Uncharacterized protein AF_1864, ssRNA | | Authors: | Yuan, Y.A, Yan, X. | | Deposit date: | 2014-09-25 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of CRISPR-associated Csx3 reveal a manganese-dependent deadenylation exoribonuclease.
Rna Biol., 12, 2015
|
|
3ZXI
 
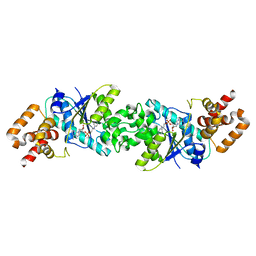 | | Crystal structure of human mitochondrial tyrosyl-tRNA synthetase in complex with a tyrosyl-adenylate analog | | Descriptor: | PHOSPHORIC ACID 2-AMINO-3-(4-HYDROXY-PHENYL)-PROPYL ESTER ADENOSIN-5'YL ESTER, TYROSYL-TRNA SYNTHETASE, MITOCHONDRIAL | | Authors: | Bonnefond, L, Frugier, M, Rudinger-Thirion, J, Balg, C, Chenevert, R, Lorber, B, Giege, R, Sauter, C. | | Deposit date: | 2011-08-11 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Exploiting Protein Engineering and Crystal Polymorphism for Successful X-Ray Structure Determination
Cryst.Growth Des., 11, 2011
|
|
3AY0
 
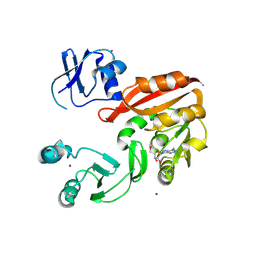 | | Crystal structure of Methanocaldococcus jannaschii Trm5 in complex with adenosine | | Descriptor: | ADENOSINE, Uncharacterized protein MJ0883, ZINC ION | | Authors: | Goto-Ito, S, Ito, T, Hou, Y.M, Yokoyama, S. | | Deposit date: | 2011-04-21 | | Release date: | 2011-08-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Differentiating analogous tRNA methyltransferases by fragments of the methyl donor.
Rna, 17, 2011
|
|
1L3Z
 
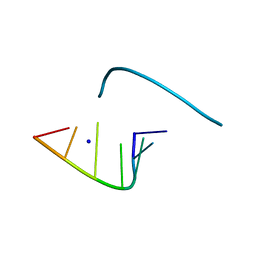 | | Crystal Structure Analysis of an RNA Heptamer | | Descriptor: | 5'-R(*GP*UP*AP*UP*AP*CP*A)-3', SODIUM ION | | Authors: | Shi, K, Pan, B, Sundaralingam, M. | | Deposit date: | 2002-03-04 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The crystal structure of an alternating RNA heptamer r(GUAUACA)
forming a six base-paired duplex with 3'-end adenine overhangs
Nucleic Acids Res., 31, 2003
|
|
1X9C
 
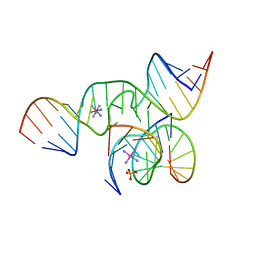 | | An all-RNA Hairpin Ribozyme with mutation U39C | | Descriptor: | 5'-R(*CP*GP*GP*UP*GP*AP*GP*AP*AP*GP*GP*G)-3', 5'-R(*GP*GP*CP*AP*GP*AP*GP*AP*AP*AP*CP*AP*CP*AP*CP*GP*A)-3', 5'-R(*UP*CP*CP*CP*(A2M)P*GP*UP*CP*CP*AP*CP*CP*G)-3', ... | | Authors: | Alam, S, Grum-Tokars, V, Krucinska, J, Kundracik, M.L, Wedekind, J.E. | | Deposit date: | 2004-08-20 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Conformational Heterogeneity at Position U37 of an All-RNA Hairpin Ribozyme with Implications for Metal Binding and the Catalytic Structure of the S-Turn.
Biochemistry, 44, 2005
|
|
1X9K
 
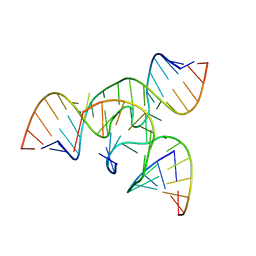 | | An all-RNA Hairpin Ribozyme with mutation U39C | | Descriptor: | 5'-R(*AP*AP*UP*AP*GP*AP*GP*AP*AP*GP*CP*GP*A)-3', 5'-R(*GP*GP*CP*AP*GP*AP*GP*AP*AP*AP*CP*AP*CP*AP*CP*GP*A)-3', 5'-R(*UP*CP*GP*CP*AP*GP*UP*CP*CP*UP*AP*UP*U)-3', ... | | Authors: | Alam, S, Grum-Tokars, V, Krucinska, J, Kundracik, M.L, Wedekind, J.E. | | Deposit date: | 2004-08-21 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Conformational Heterogeneity at Position U37 of an All-RNA Hairpin Ribozyme with Implications for Metal Binding and the Catalytic Structure of the S-Turn.
Biochemistry, 44, 2005
|
|
4AOB
 
 | | SAM-I riboswitch containing the T. solenopsae Kt-23 in complex with S- adenosyl methionine | | Descriptor: | BARIUM ION, POTASSIUM ION, S-ADENOSYLMETHIONINE, ... | | Authors: | Schroeder, K.T, Daldrop, P, McPhee, S.A, Lilley, D.M.J. | | Deposit date: | 2012-03-25 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure and Folding of a Rare, Natural Kink Turn in RNA with an Aa Pair at the 2B2N Position.
RNA, 18, 2012
|
|
1JZC
 
 | |
7S0D
 
 | |
7S0E
 
 | |
7S0B
 
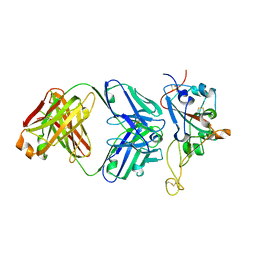 | | Structure of the SARS-CoV-2 RBD in complex with neutralizing antibody N-612-056 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-612-056 Fab Heavy Chain, N-612-056 Light Chain, ... | | Authors: | Tanaka, S, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2021-08-30 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Rapid identification of neutralizing antibodies against SARS-CoV-2 variants by mRNA display.
Cell Rep, 38, 2022
|
|
7S0C
 
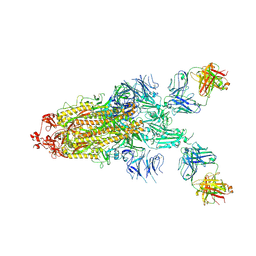 | |
3C2P
 
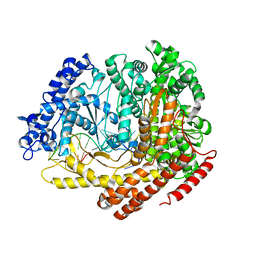 | |
3C3L
 
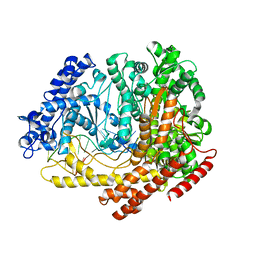 | |
3C46
 
 | |
4B5R
 
 | | SAM-I riboswitch bearing the H. marismortui K-t-7 | | Descriptor: | BARIUM ION, POTASSIUM ION, S-ADENOSYLMETHIONINE, ... | | Authors: | Daldrop, P, Lilley, D.M.J. | | Deposit date: | 2012-08-07 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The Plasticity of a Structural Motif in RNA: Structural Polymorphism of a Kink Turn as a Function of its Environment.
RNA, 19, 2013
|
|
4HVC
 
 | | Crystal structure of human prolyl-tRNA synthetase in complex with halofuginone and ATP analogue | | Descriptor: | 7-bromo-6-chloro-3-{3-[(2R,3S)-3-hydroxypiperidin-2-yl]-2-oxopropyl}quinazolin-4(3H)-one, Bifunctional glutamate/proline--tRNA ligase, MAGNESIUM ION, ... | | Authors: | Zhou, H, Sun, L, Yang, X.L, Schimmel, P. | | Deposit date: | 2012-11-06 | | Release date: | 2013-01-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ATP-directed capture of bioactive herbal-based medicine on human tRNA synthetase.
Nature, 494, 2012
|
|
1D16
 
 | |
6H62
 
 | | QTRT1, the catalytic subunit of murine tRNA-Guanine Transglycosylase | | Descriptor: | GLYCEROL, Queuine tRNA-ribosyltransferase catalytic subunit 1, ZINC ION | | Authors: | Behrens, C, Heine, A, Reuter, K. | | Deposit date: | 2018-07-26 | | Release date: | 2019-08-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.684 Å) | | Cite: | Structural and Biochemical Investigation of the Heterodimeric Murine tRNA-Guanine Transglycosylase.
Acs Chem.Biol., 17, 2022
|
|
1I2Y
 
 | | 1.66 A STRUCTURE OF A-DUPLEX WITH BULGED ADENOSINE, SPERMINE FORM | | Descriptor: | DNA/RNA (5'-R(*GP*CP*G)-D(P*AP*TP*AP*T)-R(P*AP*CP*GP*U)-3'), SPERMINE | | Authors: | Tereshko, V, Wallace, S, Usman, N, Wincott, F, Egli, M. | | Deposit date: | 2001-02-12 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | X-ray crystallographic observation of "in-line" and "adjacent" conformations in a bulged self-cleaving RNA/DNA hybrid.
RNA, 7, 2001
|
|
1I2X
 
 | | 2.4 A STRUCTURE OF A-DUPLEX WITH BULGED ADENOSINE, SPERMIDINE FORM | | Descriptor: | DNA/RNA (5'-R(*GP*CP*G)-D(P*AP*TP*AP*T)-R(P*AP*CP*GP*U)-3'), SPERMIDINE | | Authors: | Tereshko, V, Wallace, S, Usman, N, Wincott, F, Egli, M. | | Deposit date: | 2001-02-12 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystallographic observation of "in-line" and "adjacent" conformations in a bulged self-cleaving RNA/DNA hybrid.
RNA, 7, 2001
|
|
