5YG9
 
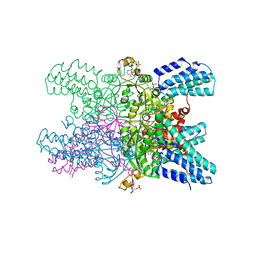 | | Crystal structure of ribose-1,5-bisphosphate isomerase mutant C135S from Pyrococcus horikoshii OT3 in complex with ribose-1,5-bisphosphate, AMP and GMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,5-di-O-phosphono-alpha-D-ribofuranose, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Gogoi, P, Kanaujia, S.P. | | Deposit date: | 2017-09-22 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A presumed homologue of the regulatory subunits of eIF2B functions as ribose-1,5-bisphosphate isomerase in Pyrococcus horikoshii OT3.
Sci Rep, 8, 2018
|
|
7KAC
 
 | | Crystal structure of HPK1 (MAP4K1) kinase in complex with 5-{[4-{[(1S)-2-HYDROXY-1-PHENYLETHYL]AMINO}-5-(1,3,4-OXADIAZOL-2-YL)PYRIMIDIN-2-YL]AMINO}-3,3-DIMETHYL-2-BENZOFURAN-1(3H)-ONE | | Descriptor: | 5-{[4-{[(1S)-2-hydroxy-1-phenylethyl]amino}-5-(1,3,4-oxadiazol-2-yl)pyrimidin-2-yl]amino}-3,3-dimethyl-2-benzofuran-1(3H)-one, Isoform 2 of Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Sheriff, S. | | Deposit date: | 2020-09-30 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Using yeast surface display to engineer a soluble and crystallizable construct of hematopoietic progenitor kinase 1 (HPK1).
Acta Crystallogr.,Sect.F, 77, 2021
|
|
4OPZ
 
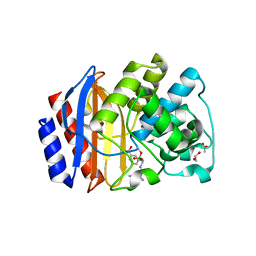 | | Crystal structure of stabilized TEM-1 beta-lactamase variant v.13 carrying G238S mutation in complex with boron-based inhibitor EC25 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(2R)-2-{[(2R)-2-amino-2-phenylacetyl]amino}-2-(dihydroxyboranyl)ethyl]benzoic acid, ACETATE ION, ... | | Authors: | Dellus-Gur, E, Elias, M, Fraser, J.S, Tawfik, D.S. | | Deposit date: | 2014-02-07 | | Release date: | 2015-05-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Negative Epistasis and Evolvability in TEM-1 beta-Lactamase--The Thin Line between an Enzyme's Conformational Freedom and Disorder.
J. Mol. Biol., 427, 2015
|
|
5VCN
 
 | | THE CRYSTAL STRUCTURE OF DER P 1 ALLERGEN COMPLEXED WITH FAB FRAGMENT OF MAB 5H8 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Osinski, T, Majorek, K.A, Pomes, A, Offermann, L.R, Osinski, S, Glesner, J, Vailes, L.D, Chapman, M.D, Minor, W, Chruszcz, M. | | Deposit date: | 2017-03-31 | | Release date: | 2017-04-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Analysis of Der p 1-Antibody Complexes and Comparison with Complexes of Proteins or Peptides with Monoclonal Antibodies.
J. Immunol., 195, 2015
|
|
4DQF
 
 | | Crystal Structure of (G16A/L38A) HIV-1 Protease in Complex with DRV | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, 1,2-ETHANEDIOL, Aspartyl protease, ... | | Authors: | Schiffer, C.A, Mittal, S. | | Deposit date: | 2012-02-15 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Hydrophobic core flexibility modulates enzyme activity in HIV-1 protease.
J.Am.Chem.Soc., 134, 2012
|
|
4DVW
 
 | | Crystal structure of clade A/E 93TH057 HIV-1 gp120 core in complex with MAE-II-167 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, N-(4-chloro-3-fluorophenyl)-N'-[(3aS,6aS)-hexahydrocyclopenta[c]pyrrol-3a(1H)-ylmethyl]ethanediamide, ... | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2012-02-23 | | Release date: | 2013-02-27 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of HIV-1 gp120 Envelope Glycoprotein in Complex with NBD Analogues That Target the CD4-Binding Site.
Plos One, 9, 2014
|
|
3NCZ
 
 | |
5HG7
 
 | | EGFR (L858R, T790M, V948R) in complex with 1-{(3R,4R)-3-[5-Chloro-2-(1-methyl-1H-pyrazol-4-ylamino)-7H-pyrrolo[2,3-d]pyrimidin-4-yloxymethyl]-4-methoxy-pyrrolidin-1-yl}propenone (PF-06459988) | | Descriptor: | 1-{(3R,4R)-3-[({5-chloro-2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-methoxypyrrolidin-1-yl}propan-1-one, Epidermal growth factor receptor, SULFATE ION | | Authors: | Gajiwala, K.S. | | Deposit date: | 2016-01-08 | | Release date: | 2016-01-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of 1-{(3R,4R)-3-[({5-Chloro-2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-methoxypyrrolidin-1-yl}prop-2-en-1-one (PF-06459988), a Potent, WT Sparing, Irreversible Inhibitor of T790M-Containing EGFR Mutants.
J.Med.Chem., 59, 2016
|
|
5ZGW
 
 | |
5HG9
 
 | | EGFR (L858R, T790M, V948R) in complex with 1-[(3R,4R)-3-[({2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-(trifluoromethyl)pyrrolidin-1-yl]prop-2-en-1-one | | Descriptor: | 1-[(3R,4R)-3-[({2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-(trifluoromethyl)pyrr olidin-1-yl]propan-1-one, Epidermal growth factor receptor, GLYCEROL, ... | | Authors: | Gajiwala, K.S. | | Deposit date: | 2016-01-08 | | Release date: | 2016-02-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of 1-{(3R,4R)-3-[({5-Chloro-2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-methoxypyrrolidin-1-yl}prop-2-en-1-one (PF-06459988), a Potent, WT Sparing, Irreversible Inhibitor of T790M-Containing EGFR Mutants.
J.Med.Chem., 59, 2016
|
|
2XON
 
 | | Structure of TmCBM61 in complex with beta-1,4-galactotriose at 1.4 A resolution | | Descriptor: | 1,2-ETHANEDIOL, ARABINOGALACTAN ENDO-1,4-BETA-GALACTOSIDASE, CALCIUM ION, ... | | Authors: | Cid, M, Lodberg-Pedersen, H, Kaneko, S, Coutinho, P.M, Henrissat, B, Willats, W.G.T, Boraston, A.B. | | Deposit date: | 2010-08-20 | | Release date: | 2010-09-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Recognition of the Helical Structure of Beta-1,4-Galactan by a New Family of Carbohydrate-Binding Modules.
J.Biol.Chem., 285, 2010
|
|
5CJF
 
 | | The crystal structure of the human carbonic anhydrase XIV in complex with a 1,1'-biphenyl-4-sulfonamide inhibitor. | | Descriptor: | 4'-(4-aminobenzoyl)biphenyl-4-sulfonamide, Carbonic anhydrase 14, GLYCEROL, ... | | Authors: | Alterio, V, De Simone, G. | | Deposit date: | 2015-07-14 | | Release date: | 2015-11-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of 1,1'-Biphenyl-4-sulfonamides as a New Class of Potent and Selective Carbonic Anhydrase XIV Inhibitors.
J.Med.Chem., 58, 2015
|
|
5OUJ
 
 | | Crystal structure of human AKR1B1 complexed with NADP+ and compound 39 | | Descriptor: | 2-[(1~{R})-5-(4-chlorophenyl)-9-fluoranyl-3-methyl-1-oxidanyl-1~{H}-pyrimido[4,5-c]quinolin-2-yl]ethanoic acid, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Metwally, K, Podjarny, A. | | Deposit date: | 2017-08-24 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Design, synthesis, structure-activity relationships and X-ray structural studies of novel 1-oxopyrimido[4,5-c]quinoline-2-acetic acid derivatives as selective and potent inhibitors of human aldose reductase.
Eur J Med Chem, 152, 2018
|
|
6LJ6
 
 | |
2XIX
 
 | | Protein kinase Pim-1 in complex with fragment-1 from crystallographic fragment screen | | Descriptor: | 3,5-DIAMINO-1H-[1,2,4]TRIAZOLE, PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2XJ2
 
 | | Protein kinase Pim-1 in complex with small molecule inhibitor | | Descriptor: | (2E)-3-{3-[6-(4-methyl-1,4-diazepan-1-yl)pyrazin-2-yl]phenyl}prop-2-enoic acid, PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
4BGV
 
 | |
2XIY
 
 | | Protein kinase Pim-1 in complex with fragment-2 from crystallographic fragment screen | | Descriptor: | 2-HYDROXYMETHYL-BENZOIMIDAZOLE, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3NIY
 
 | | Crystal structure of native xylanase 10B from Thermotoga petrophila RKU-1 | | Descriptor: | ACETATE ION, Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Santos, C.R, Meza, A.N, Trindade, D.M, Ruller, R, Squina, F.M, Prade, R.A, Murakami, M.T. | | Deposit date: | 2010-06-16 | | Release date: | 2011-05-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Thermal-induced conformational changes in the product release area drive the enzymatic activity of xylanases 10B: Crystal structure, conformational stability and functional characterization of the xylanase 10B from Thermotoga petrophila RKU-1.
Biochem.Biophys.Res.Commun., 403, 2010
|
|
4QAG
 
 | | Structure of a dihydroxycoumarin active-site inhibitor in complex with the RNASE H domain of HIV-1 reverse transcriptase | | Descriptor: | (7,8-dihydroxy-2-oxo-2H-chromen-4-yl)acetic acid, MANGANESE (II) ION, Reverse transcriptase/ribonuclease H | | Authors: | Himmel, D.M, Ho, W.C, Arnold, E. | | Deposit date: | 2014-05-04 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.712 Å) | | Cite: | Structure of a Dihydroxycoumarin Active-Site Inhibitor in Complex with the RNase H Domain of HIV-1 Reverse Transcriptase and Structure-Activity Analysis of Inhibitor Analogs.
J.Mol.Biol., 426, 2014
|
|
1HT6
 
 | | CRYSTAL STRUCTURE AT 1.5A RESOLUTION OF THE BARLEY ALPHA-AMYLASE ISOZYME 1 | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-AMYLASE ISOZYME 1, CALCIUM ION | | Authors: | Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2000-12-29 | | Release date: | 2003-07-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of barley alpha-amylase isozyme 1 reveals a novel role of domain C in substrate recognition and binding: a pair of sugar tongs
Structure, 11, 2003
|
|
6LZO
 
 | | Thermolysin with 1,10-phenanthroline | | Descriptor: | 1,10-PHENANTHROLINE, CALCIUM ION, Thermolysin | | Authors: | Nam, K.H. | | Deposit date: | 2020-02-19 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of metal chelation of the metalloproteinase thermolysin by 1,10-phenanthroline.
J.Inorg.Biochem., 215, 2021
|
|
4S0Z
 
 | | Crystal structure of M26V human DJ-1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Protein DJ-1 | | Authors: | Milkovic, N.M, Wilson, M.A. | | Deposit date: | 2015-01-07 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Transient sampling of aggregation-prone conformations causes pathogenic instability of a parkinsonian mutant of DJ-1 at physiological temperature.
Protein Sci., 24, 2015
|
|
3ANY
 
 | |
1TJG
 
 | | Crystal Structure of the broadly neutralizing anti-HIV-1 antibody 2F5 in complex with a gp41 7mer epitope | | Descriptor: | 1,2-ETHANEDIOL, Envelope glycoprotein GP41, FAB 2F5 Heavy Chain, ... | | Authors: | Ofek, G, Tang, M, Sambor, A, Katinger, H, Mascola, J.R, Wyatt, R, Kwong, P.D. | | Deposit date: | 2004-06-04 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanistic analysis of the Anti-Human Immunodeficiency Virus Type 1 antibody 2F5 in complex with its gp41 epitope
J.Virol., 78, 2004
|
|
