2WPU
 
 | | Chaperoned ruthenium metallodrugs that recognize telomeric DNA | | Descriptor: | (3AS,4S,6AR)-4-(5-((3R,4R)-3,4-DIAMINOPYRROLIDIN-1-YL)-5-OXOPENTYL)TETRAHYDRO-1H-THIENO[3,4-D]IMIDAZOL-2(3H)-ONE-P-CYMENE-CHLORO-RUTHENIUM(III), GLYCEROL, STREPTAVIDIN, ... | | Authors: | Heinisch, T, Schirmer, T, Zimbron, J.M, Sardo, A, Wohlschlager, T, Gradinaru, J, Creus, M, Ward, T.R. | | Deposit date: | 2009-08-10 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Chemo-Genetic Optimization of DNA Recognition by Metallodrugs Using a Presenter-Protein Strategy.
Chemistry, 16, 2010
|
|
1TLO
 
 | | High resolution crystal structure of calpain I protease core in complex with E64 | | Descriptor: | CALCIUM ION, Calpain 1, large [catalytic] subunit, ... | | Authors: | Moldoveanu, T, Campbell, R.L, Cuerrier, D, Davies, P.L. | | Deposit date: | 2004-06-09 | | Release date: | 2004-11-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Calpain-E64 and -Leupeptin Inhibitor Complexes Reveal Mobile Loops Gating the Active Site
J.Mol.Biol., 343, 2004
|
|
8VEK
 
 | | IsPETase - ACC mutant | | Descriptor: | GLYCEROL, Poly(ethylene terephthalate) hydrolase, SULFATE ION | | Authors: | Joho, Y, Royan, S, Newton, S, Caputo, A.T, Ardevol Grau, A, Jackson, C. | | Deposit date: | 2023-12-19 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Enhancing PET Degrading Enzymes: A Combinatory Approach.
Chembiochem, 25, 2024
|
|
8V0R
 
 | | SARS-CoV-2 Omicron-XBB.1.5 3-RBD down Spike Protein Trimer 1 (S-GSAS-Omicron-XBB.1.5) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, Q.E, Acharya, P. | | Deposit date: | 2023-11-17 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition.
Mol.Cell, 84, 2024
|
|
8VEL
 
 | | IsPETase - ACCCC mutant | | Descriptor: | Poly(ethylene terephthalate) hydrolase, SULFATE ION | | Authors: | Joho, Y, Royan, S, Newton, S, Caputo, A.T, Ardevol Grau, A, Jackson, C. | | Deposit date: | 2023-12-19 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.624 Å) | | Cite: | Enhancing PET Degrading Enzymes: A Combinatory Approach.
Chembiochem, 25, 2024
|
|
1XQO
 
 | | Crystal structure of native Pa-AGOG, 8-oxoguanine DNA glycosylase from Pyrobaculum aerophilum | | Descriptor: | 8-oxoguanine DNA glycosylase | | Authors: | Lingaraju, G.M, Sartori, A.A, Kostrewa, D, Prota, A.E, Jiricny, J, Winkler, F.K. | | Deposit date: | 2004-10-13 | | Release date: | 2004-11-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | A DNA glycosylase from Pyrobaculum aerophilum with an 8-oxoguanine binding mode and a noncanonical helix-hairpin-helix structure
Structure, 13, 2005
|
|
1LL4
 
 | | STRUCTURE OF C. IMMITIS CHITINASE 1 COMPLEXED WITH ALLOSAMIDIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, CHITINASE 1 | | Authors: | Bortone, K, Monzingo, A.F, Ernst, S, Robertus, J.D. | | Deposit date: | 2002-04-26 | | Release date: | 2002-09-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | THE STRUCTURE OF AN ALLOSAMIDIN COMPLEX WITH THE Coccidioides IMMITIS CHITINASE DEFINES A ROLE FOR A SECOND ACID RESIDUE IN SUBSTRATE-ASSISTED MECHANISM
J.Mol.Biol., 320, 2002
|
|
8VLU
 
 | | Cryo-EM structure of human HGSNAT bound with CoA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COENZYME A, Heparan-alpha-glucosaminide N-acetyltransferase | | Authors: | Li, F, Zhao, B. | | Deposit date: | 2024-01-12 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structural and mechanistic insights into a lysosomal membrane enzyme HGSNAT involved in Sanfilippo syndrome.
Nat Commun, 15, 2024
|
|
7CH6
 
 | | Cryo-EM structure of E.coli MlaFEB with AMPPNP | | Descriptor: | Lipid asymmetry maintenance ABC transporter permease subunit MlaE, Lipid asymmetry maintenance protein MlaB, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Zhou, C, Shi, H, Zhang, M, Huang, Y. | | Deposit date: | 2020-07-05 | | Release date: | 2021-08-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Insight into Phospholipid Transport by the MlaFEBD Complex from P. aeruginosa.
J.Mol.Biol., 433, 2021
|
|
1TH0
 
 | | Structure of human Senp2 | | Descriptor: | Sentrin-specific protease 2 | | Authors: | Reverter, D, Lima, C.D. | | Deposit date: | 2004-05-31 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A basis for SUMO protease specificity provided by analysis of human Senp2 and a
Senp2-SUMO complex
Structure, 12, 2004
|
|
1TH9
 
 | | Effect of Shuttle Location and pH Environment on H+ Transfer in Human Carbonic Anhydrase II | | Descriptor: | Carbonic anhydrase II, SULFATE ION, ZINC ION | | Authors: | Fisher, Z, Hernandez Prada, J.A, Tu, C.K, Duda, D, Yoshioka, C, An, H, Govindasamy, L, Silverman, D.N, McKenna, R. | | Deposit date: | 2004-06-01 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural and Kinetic Characterization of Active-Site Histidine as a Proton Shuttle in Catalysis by Human Carbonic Anhydrase II
Biochemistry, 44, 2005
|
|
2WXT
 
 | | Clostridium perfringens alpha-toxin strain NCTC8237 | | Descriptor: | CADMIUM ION, CALCIUM ION, PHOSPHOLIPASE C, ... | | Authors: | Justin, N, Naylor, C.E, Basak, A.K. | | Deposit date: | 2009-11-10 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Comparison of a Nontoxic Variant of Clostridium Perfringens [Alpha]-Toxin with the Toxic Wild-Type Strain
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1TK6
 
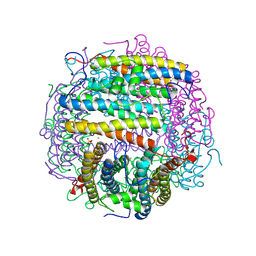 | | Iron-oxo clusters biomineralizing on protein surfaces. Structural analysis of H.salinarum DpsA in its low and high iron states | | Descriptor: | FE (III) ION, Iron-rich dpsA-homolog protein, MAGNESIUM ION, ... | | Authors: | Zeth, K, Offermann, S, Essen, L.O, Oesterhelt, D. | | Deposit date: | 2004-06-08 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Iron-oxo clusters biomineralizing on protein surfaces: structural analysis of Halobacterium salinarum DpsA in its low- and high-iron states.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1LO7
 
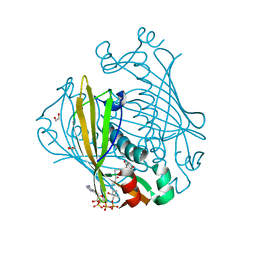 | | X-ray structure of 4-Hydroxybenzoyl CoA Thioesterase complexed with 4-hydroxyphenacyl CoA | | Descriptor: | 1,2-ETHANEDIOL, 4-HYDROXYPHENACYL COENZYME A, 4-hydroxybenzoyl-CoA Thioesterase | | Authors: | Thoden, J.B, Holden, H.M, Zhuang, Z, Dunaway-Mariano, D. | | Deposit date: | 2002-05-06 | | Release date: | 2002-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystallographic analyses of inhibitor and substrate complexes of wild-type and mutant 4-hydroxybenzoyl-CoA thioesterase.
J.Biol.Chem., 277, 2002
|
|
1XX8
 
 | |
1SU5
 
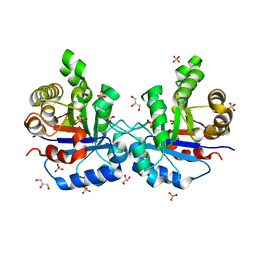 | | Understanding protein lids: Structural analysis of active hinge mutants in triosephosphate isomerase | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-26 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
1XXJ
 
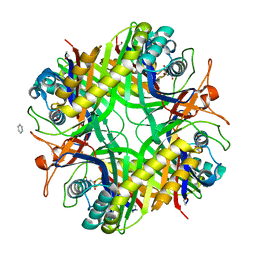 | | Urate oxidase from aspergillus flavus complexed with 5-amino 6-nitro uracil | | Descriptor: | 5-AMINO-6-NITROPYRIMIDINE-2,4(1H,3H)-DIONE, BENZENE, Uricase | | Authors: | Retailleau, P, Colloc'h, N, Vivares, D, Bonnete, F, Castro, B, El Hajji, M, Prange, T. | | Deposit date: | 2004-11-05 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Urate oxidase from Aspergillus flavus: new crystal-packing contacts in relation to the content of the active site.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
8V0Q
 
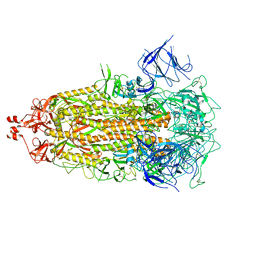 | | SARS-CoV-2 Omicron-XBB.1.16 3-RBD down Spike Protein Trimer 3 (S-GSAS-Omicron-XBB.1.16) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, Q.E, Acharya, P. | | Deposit date: | 2023-11-17 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition.
Mol.Cell, 84, 2024
|
|
8V0U
 
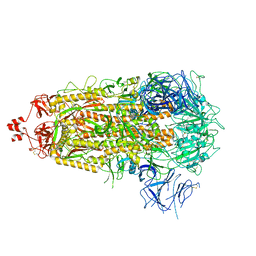 | | SARS-CoV-2 Omicron-XBB.1.5 3-RBD down Spike Protein Trimer 4 (S-GSAS-Omicron-XBB.1.5) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, Q.E, Acharya, P. | | Deposit date: | 2023-11-17 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition.
Mol.Cell, 84, 2024
|
|
7CQB
 
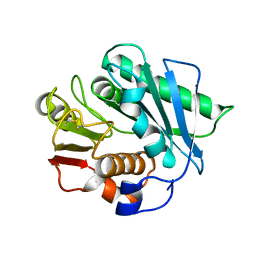 | |
1SW7
 
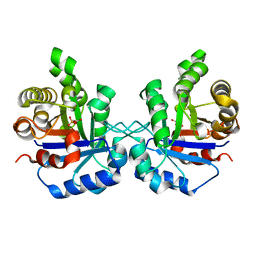 | | Triosephosphate isomerase from Gallus gallus, loop 6 mutant K174N, T175S, A176S | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate isomerase | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-30 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
1XFA
 
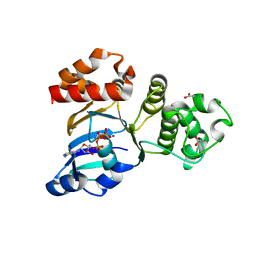 | | Structure of NBD1 from murine CFTR- F508R mutant | | Descriptor: | ACETIC ACID, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Thibodeau, P.H, Brautigam, C.A, Machius, M, Thomas, P.J. | | Deposit date: | 2004-09-14 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Side chain and backbone contributions of Phe508 to CFTR folding.
Nat.Struct.Mol.Biol., 12, 2005
|
|
8V7Z
 
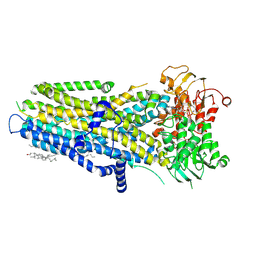 | | Phosphorylated, ATP-bound, E1371Q human cystic fibrosis transmembrane conductance regulator (E1371Q-CFTR) | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ADENOSINE-5'-TRIPHOSPHATE, CHOLESTEROL, ... | | Authors: | Gao, X, Hwang, T. | | Deposit date: | 2023-12-04 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Allosteric inhibition of CFTR gating by CFTRinh-172 binding in the pore.
Nat Commun, 15, 2024
|
|
7CUQ
 
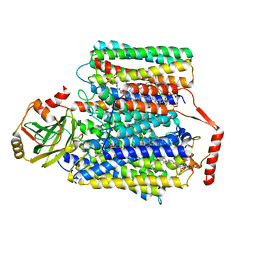 | | 2.55-Angstrom Cryo-EM structure of Cytochrome bo3 from Escherichia coli in Native Membrane | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Li, J, Han, L, Gennis, R.B, Zhu, J.P, Zhang, K. | | Deposit date: | 2020-08-24 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM structures of Escherichia coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2VS3
 
 | | THE BINDING OF UDP-GALACTOSE BY AN ACTIVE SITE MUTANT OF alpha-1,3 GALACTOSYLTRANSFERASE (alpha3GT) | | Descriptor: | MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Tumbale, P, Jamaluddin, H, Thiyagarajan, N, Brew, K, Acharya, K.R. | | Deposit date: | 2008-04-18 | | Release date: | 2008-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Udp-Galactose Binding by Alpha- 1,3-Galactosyltransferase (Alpha3Gt): Role of Negative Charge on Aspartic Acid 316 in Structure and Activity.
Biochemistry, 47, 2008
|
|
