4WEA
 
 | | Structure and receptor binding prefereneces of recombinant human A(H3N2) virus hemagglutinins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Guo, Z, Villanueva, J.M, Stevens, J. | | Deposit date: | 2014-09-09 | | Release date: | 2015-02-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and receptor binding preferences of recombinant human A(H3N2) virus hemagglutinins.
Virology, 477C, 2015
|
|
3E6L
 
 | | Structure of murine INOS oxygenase domain with inhibitor AR-C132283 | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ETHYL 4-[(4-CHLOROPYRIDIN-2-YL)AMINO]PIPERIDINE-1-CARBOXYLATE, Nitric oxide synthase, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stueh, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-15 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
4WEI
 
 | | Crystal structure of the F4 fimbrial adhesin FaeG in complex with lactose | | Descriptor: | K88 fimbrial protein AD, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Moonens, K, Van den Broeck, I, De Kerpel, M, Deboeck, F, Raymaekers, H, Remaut, H, De Greve, H. | | Deposit date: | 2014-09-10 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Functional Insight into the Carbohydrate Receptor Binding of F4 Fimbriae-producing Enterotoxigenic Escherichia coli.
J.Biol.Chem., 290, 2015
|
|
3UVP
 
 | | Human p38 MAP Kinase in Complex with a Benzamide Substituted Benzosuberone | | Descriptor: | Mitogen-activated protein kinase 14, N-{2-fluoro-5-[(5-oxo-6,7,8,9-tetrahydro-5H-benzo[7]annulen-2-yl)amino]phenyl}benzamide, octyl beta-D-glucopyranoside | | Authors: | Mayer-Wrangowski, S.C, Richters, A, Gruetter, C, Rauh, D. | | Deposit date: | 2011-11-30 | | Release date: | 2012-11-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting the Hinge Glycine Flip and the Activation Loop: Novel Approach to Potent p38 alpha Inhibitors.
J.Med.Chem., 55, 2012
|
|
6SS7
 
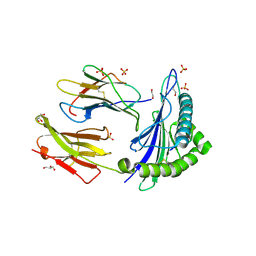 | | Human Leukocyte Antigen Class I A02 Carrying LLWAGPMAV | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Rizkallah, P.J, Bovay, A. | | Deposit date: | 2019-09-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a superagonist variant of the immunodominant Yellow fever virus epitope NS4b214-222by combinatorial peptide library screening.
Mol.Immunol., 125, 2020
|
|
4UTC
 
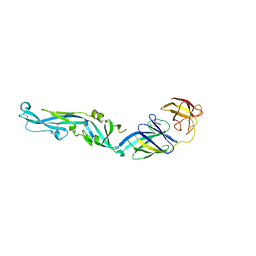 | | Crystal structure of dengue 2 virus envelope glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ENVELOPE GLYCOPROTEIN E, ... | | Authors: | Kikuti, C, Rouvinski, A, Guardado-Calvo, P, Barba-Spaeth, G, Duquerroy, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2014-07-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Recognition Determinants of Broadly Neutralizing Human Antibodies Against Dengue Viruses.
Nature, 520, 2015
|
|
4UV7
 
 | | The complex structure of extracellular domain of EGFR and GC1118A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, EPIDERMAL GROWTH FACTOR RECEPTOR, ... | | Authors: | Yoo, J.H, Cho, H.S. | | Deposit date: | 2014-08-05 | | Release date: | 2015-10-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Gc1118, an Anti-Egfr Antibody with a Distinct Binding Epitope and Superior Inhibitory Activity Against High-Affinity Egfr Ligands.
Mol.Cancer Ther., 15, 2016
|
|
6SXW
 
 | | Crystal structure of the first RRM domain of human Zinc finger protein 638 (ZNF638) | | Descriptor: | SULFATE ION, Zinc finger protein 638 | | Authors: | Newman, J.A, Aitkenhead, H, Wang, D, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-09-26 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | Crystal structure of the first RRM domain of human Zinc finger protein 638 (ZNF638)
To Be Published
|
|
4UYW
 
 | |
4UZI
 
 | | Crystal Structure of AauDyP Complexed with Imidazole | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Strittmatter, E, Liers, C, Ullrich, R, Hofrichter, M, Piontek, K, Plattner, D.A. | | Deposit date: | 2014-09-05 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Toolbox of Auricularia Auricula-Judae Dye-Decolorizing Peroxidase - Identification of Three New Potential Substrate-Interaction Sites.
Arch.Biochem.Biophys., 574, 2015
|
|
6NEA
 
 | | Human Acetylcholinesterase in complex with reactivator, HLo7 | | Descriptor: | 1-[({2,4-BIS[(E)-(HYDROXYIMINO)METHYL]PYRIDINIUM-1-YL}METHOXY)METHYL]-4-CARBAMOYLPYRIDINIUM, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bester, S.M, McGuire, J, Height, J.J, Pegan, S.D. | | Deposit date: | 2018-12-17 | | Release date: | 2019-06-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.419 Å) | | Cite: | Synthesis and Molecular Properties of Nerve Agent Reactivator HLo-7 Dimethanesulfonate.
Acs Med.Chem.Lett., 10, 2019
|
|
4V0Z
 
 | |
6NFC
 
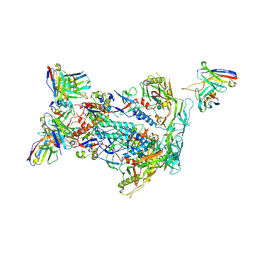 | | BG505 MD64 N332-GT5 SOSIP trimer in complex with BG18-like precursor HMP42 fragmentantigen binding and base-binding RM20A3 fragment antigen binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG18-like precursor HMP42 fragment antigen binding heavy chain, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2018-12-19 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | A generalized HIV vaccine design strategy for priming of broadly neutralizing antibody responses.
Science, 366, 2019
|
|
4MZA
 
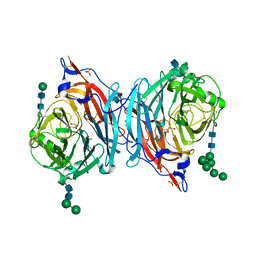 | | Crystal structure of hPIV3 hemagglutinin-neuraminidase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Xu, R, Wilson, I.A. | | Deposit date: | 2013-09-29 | | Release date: | 2013-11-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Interaction between the hemagglutinin-neuraminidase and fusion glycoproteins of human parainfluenza virus type III regulates viral growth in vivo.
MBio, 4, 2013
|
|
4N38
 
 | | Structure of langerin CRD I313 D288 complexed with GlcNAc-beta1-3Gal-beta1-4GlcNAc-beta-CH2CH2N3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose, C-type lectin domain family 4 member K, CALCIUM ION, ... | | Authors: | Feinberg, H, Rowntree, T.J.W, Tan, S.L.W, Drickamer, K, Weis, W.I, Taylor, M.E. | | Deposit date: | 2013-10-06 | | Release date: | 2013-11-27 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Common polymorphisms in human langerin change specificity for glycan ligands.
J.Biol.Chem., 288, 2013
|
|
3M73
 
 | |
3M7B
 
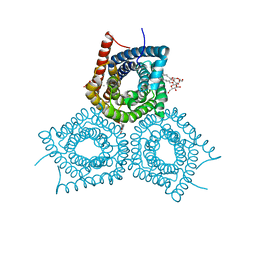 | |
4N35
 
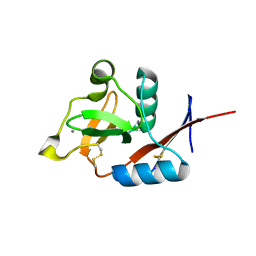 | | Structure of langerin CRD I313 complexed with GlcNAc-beta1-3Gal-beta1-4Glc-beta-CH2CH2N3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, C-type lectin domain family 4 member K, ... | | Authors: | Feinberg, H, Rowntree, T.J.W, Tan, S.L.W, Drickamer, K, Weis, W.I, Taylor, M.E. | | Deposit date: | 2013-10-06 | | Release date: | 2013-11-20 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Common polymorphisms in human langerin change specificity for glycan ligands.
J.Biol.Chem., 288, 2013
|
|
6NTL
 
 | | Crystal Structure of Recombinant Human Acetylcholinesterase Inhibited by A-234 | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Bester, S.M, Guelta, M.A, Height, J.J, Pegan, S.D. | | Deposit date: | 2019-01-29 | | Release date: | 2020-07-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Insights into inhibition of human acetylcholinesterase by Novichok, A-series Nerve Agents
To Be Published
|
|
7A4Y
 
 | | Crystal structure of the P5P6 coiled-coil in complex with nanobody Nb34. | | Descriptor: | 1,2-ETHANEDIOL, Coiled-coil P5 peptide, Coiled-coil P6 peptide, ... | | Authors: | Hadzi, S. | | Deposit date: | 2020-08-20 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.157 Å) | | Cite: | A nanobody toolbox targeting dimeric coiled-coil modules for functionalization of designed protein origami structures.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6ZTB
 
 | | Crystal Structure of human P-Cadherin EC1_EC2 | | Descriptor: | CALCIUM ION, Cadherin-3, SODIUM ION | | Authors: | Rondeau, J.M, Lehmann, S. | | Deposit date: | 2020-07-17 | | Release date: | 2021-05-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | PCA062, a P-cadherin Targeting Antibody-Drug Conjugate, Displays Potent Antitumor Activity Against P-cadherin-expressing Malignancies.
Mol.Cancer Ther., 20, 2021
|
|
4UT5
 
 | | Crystal structure of the LecB lectin from Pseudomonas aeruginosa strain PA7 in complex with lewis a tetrasaccharide | | Descriptor: | CALCIUM ION, LECB LECTIN, beta-D-galactopyranose, ... | | Authors: | Boukerb, A.M, Decor, A, Tabaroni, R, Varrot, A, Debentzmann, S, Vidal, S, Imberty, A, Cournoyer, B. | | Deposit date: | 2014-07-18 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Genomic Rearrangements and Functional Diversification of Leca and Lecb Lectin-Coding Regions Impacting the Efficacy of Glycomimetics Directed Against Pseudomonas Aeruginosa.
Front.Microbiol., 7, 2016
|
|
7A4T
 
 | | Crystal structure of the GCN coiled-coil in complex with nanobody Nb39 | | Descriptor: | 1,2-ETHANEDIOL, ACETYL GROUP, GCN4 isoform 1, ... | | Authors: | Hadzi, S. | | Deposit date: | 2020-08-20 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.124 Å) | | Cite: | A nanobody toolbox targeting dimeric coiled-coil modules for functionalization of designed protein origami structures.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7A50
 
 | | Crystal structure of the APH coiled-coil in complex with nanobody Nb26 | | Descriptor: | 1,2-ETHANEDIOL, Coiled-coil APH, Nanobody Nb26 | | Authors: | Hadzi, S. | | Deposit date: | 2020-08-20 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | A nanobody toolbox targeting dimeric coiled-coil modules for functionalization of designed protein origami structures.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6ZTF
 
 | | Crystal Structure of the anti-human P-Cadherin Fab CQY684 | | Descriptor: | CQY684 Fab heavy-chain, CQY684 Fab light-chain | | Authors: | Rondeau, J.M, Lehmann, S. | | Deposit date: | 2020-07-20 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | PCA062, a P-cadherin Targeting Antibody-Drug Conjugate, Displays Potent Antitumor Activity Against P-cadherin-expressing Malignancies.
Mol.Cancer Ther., 20, 2021
|
|
