1D91
 
 | |
1LWB
 
 | |
1W1Q
 
 | | Plant Cytokinin Dehydrogenase in Complex with Isopentenyladenine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CYTOKININ DEHYDROGENASE 1, ... | | Authors: | Malito, E, Mattevi, A. | | Deposit date: | 2004-06-23 | | Release date: | 2004-08-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of Michaelis and Product Complexes of Plant Cytokinin Dehydrogenase: Implications for Flavoenzyme Catalysis
J.Mol.Biol., 341, 2004
|
|
3R3Q
 
 | | Crystal structure of the yeast Vps23 UEV domain | | Descriptor: | ACETATE ION, CHLORIDE ION, IMIDAZOLE, ... | | Authors: | Ren, X, Hurley, J.H. | | Deposit date: | 2011-03-16 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis for endosomal recruitment of ESCRT-I by ESCRT-0 in yeast.
Embo J., 30, 2011
|
|
3CXF
 
 | | Crystal structure of transthyretin variant Y114H | | Descriptor: | Transthyretin | | Authors: | Cendron, L, Zanotti, G, Folli, C, Alfieri, B, Pasquato, N, Berni, R. | | Deposit date: | 2008-04-24 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mutational analyses of protein-protein interactions between transthyretin and retinol-binding protein.
Febs J., 275, 2008
|
|
1M26
 
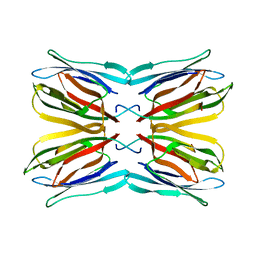 | | Crystal structure of jacalin-T-antigen complex | | Descriptor: | Jacalin, alpha chain, beta chain, ... | | Authors: | Jeyaprakash, A.A, Rani, P.G, Reddy, G.B, Banumathi, S, Betzel, C, Surolia, A, Vijayan, M. | | Deposit date: | 2002-06-21 | | Release date: | 2002-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure of the jacalin-T-antigen complex and a
comparative study of lectin-T-antigen complexs
J.Mol.Biol., 321, 2002
|
|
3TDL
 
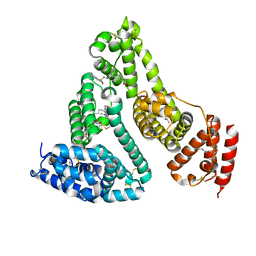 | | Structure of human serum albumin in complex with DAUDA | | Descriptor: | 11-({[5-(dimethylamino)naphthalen-1-yl]sulfonyl}amino)undecanoic acid, MYRISTIC ACID, Serum albumin | | Authors: | Wang, Y, Luo, Z, Shi, X, Wang, H, Nie, L. | | Deposit date: | 2011-08-11 | | Release date: | 2012-06-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A fluorescent fatty acid probe, DAUDA, selectively displaces two myristates bound in human serum albumin
Protein Sci., 20, 2011
|
|
1LXY
 
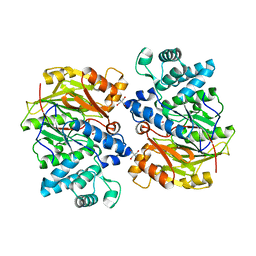 | | Crystal Structure of Arginine Deiminase covalently linked with L-citrulline | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Arginine Deiminase, CITRULLINE | | Authors: | Das, K, Buttler, G.H, Kwiatkowski, V, Yadav, P, Arnold, E. | | Deposit date: | 2002-06-06 | | Release date: | 2004-01-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of arginine deiminase with covalent reaction intermediates; implications for catalytic mechanism
Structure, 12, 2004
|
|
3CFV
 
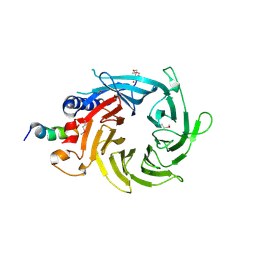 | | Structural basis of the interaction of RbAp46/RbAp48 with histone H4 | | Descriptor: | ARSENIC, Histone H4 peptide, Histone-binding protein RBBP7 | | Authors: | Pei, X.-Y, Murzina, N.V, Zhang, W, McLaughlin, S, Verreault, A, Luisi, B.F, Laue, E.D. | | Deposit date: | 2008-03-04 | | Release date: | 2008-06-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for the Recognition of Histone H4 by the Histone-Chaperone RbAp46.
Structure, 16, 2008
|
|
1MI8
 
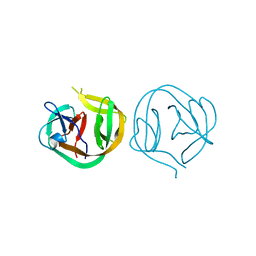 | | 2.0 Angstrom crystal structure of a DnaB intein from Synechocystis sp. PCC 6803 | | Descriptor: | DnaB intein | | Authors: | Ding, Y, Chen, X, Ferrandon, S, Xu, M, Rao, Z. | | Deposit date: | 2002-08-22 | | Release date: | 2003-08-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of mini-intein reveals a conserved catalytic module involved in side chain cyclization of asparagine during protein splicing
J.Biol.Chem., 278, 2003
|
|
1MKC
 
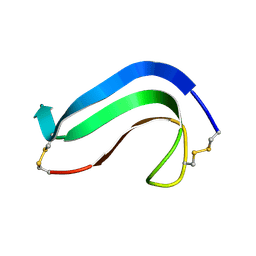 | | C-TERMINAL DOMAIN OF MIDKINE | | Descriptor: | PROTEIN (MIDKINE) | | Authors: | Iwasaki, W, Nagata, K, Hatanaka, H, Ogura, K, Inui, T, Kimura, T, Muramatsu, T, Yoshida, K, Tasumi, M, Inagaki, F. | | Deposit date: | 1999-03-16 | | Release date: | 1999-03-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of midkine, a new heparin-binding growth factor.
EMBO J., 16, 1997
|
|
3TCJ
 
 | |
1STM
 
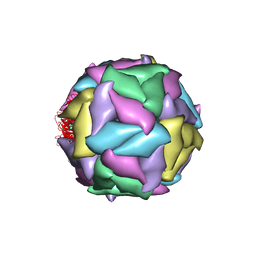 | | SATELLITE PANICUM MOSAIC VIRUS | | Descriptor: | SATELLITE PANICUM MOSAIC VIRUS | | Authors: | Ban, N, McPherson, A. | | Deposit date: | 1995-07-12 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of satellite panicum mosaic virus at 1.9 A resolution.
Nat.Struct.Biol., 2, 1995
|
|
1DUC
 
 | | EIAV DUTPASE DUDP/STRONTIUM COMPLEX | | Descriptor: | DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE, DEOXYURIDINE-5'-DIPHOSPHATE, STRONTIUM ION | | Authors: | Dauter, Z, Persson, R, Rosengren, A.M, Nyman, P.O, Wilson, K.S, Cedergren-Zeppezauer, E.S. | | Deposit date: | 1997-11-29 | | Release date: | 1998-06-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of dUTPase from equine infectious anaemia virus; active site metal binding in a substrate analogue complex.
J.Mol.Biol., 285, 1999
|
|
1T41
 
 | | Crystal structure of human aldose reductase complexed with NADP and IDD552 | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [5-FLUORO-2-({[(4,5,7-TRIFLUORO-1,3-BENZOTHIAZOL-2-YL)METHYL]AMINO}CARBONYL)PHENOXY]ACETIC ACID | | Authors: | Ruiz, F, Hazemann, I, Mitschler, A, Chevrier, B, Schneider, T, Joachimiak, A, Karplus, M, Podjarny, A. | | Deposit date: | 2004-04-28 | | Release date: | 2004-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | The crystallographic structure of the aldose reductase-IDD552 complex shows direct proton donation from tyrosine 48.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3THK
 
 | | Structure of SH3 chimera with a type II ligand linked to the chain C-terminal | | Descriptor: | BETA-MERCAPTOETHANOL, Proline-rich peptide, SULFATE ION, ... | | Authors: | Gabdulkhakov, A.G, Gushchina, L.V, Nikulin, A.D, Nikonov, S.V, Filimonov, V.V. | | Deposit date: | 2011-08-19 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-Resolution Crystal Structure of Spectrin SH3 Domain Fused with a Proline-Rich Peptide.
J.Biomol.Struct.Dyn., 29, 2011
|
|
1DV0
 
 | |
1M54
 
 | | CYSTATHIONINE-BETA SYNTHASE: REDUCED VICINAL THIOLS | | Descriptor: | CYSTATHIONINE BETA-SYNTHASE, PROTOPORPHYRIN IX CONTAINING FE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Taoka, S, Lepore, B.W, Kabil, O, Ojha, S, Ringe, D, Banerjee, R. | | Deposit date: | 2002-07-08 | | Release date: | 2002-08-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | HUMAN CYSTATHIONINE BETA-SYNTHASE IS A HEME SENSOR PROTEIN. EVIDENCE THAT THE
REDOX SENSOR IS HEME AND NOT THE VICINAL CYSTEINES IN THE CXXC MOTIF SEEN IN THE CRYSTAL STRUCTURE OF THE TRUNCATED ENZYME
BIOCHEMISTRY, 41, 2002
|
|
3ADE
 
 | | Crystal Structure of Keap1 in Complex with Sequestosome-1/p62 | | Descriptor: | Kelch-like ECH-associated protein 1, SULFATE ION, Sequestosome-1 | | Authors: | Kurokawa, H, Yamamoto, M. | | Deposit date: | 2010-01-19 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1
Nat.Cell Biol., 12, 2010
|
|
3TLU
 
 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-24' oxidized mutant in a locally-closed conformation (LC1 subtype) | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-08-30 | | Release date: | 2012-05-16 | | Last modified: | 2012-06-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
1MAP
 
 | |
3AJM
 
 | | Crystal structure of programmed cell death 10 in complex with inositol 1,3,4,5-tetrakisphosphate | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, Programmed cell death protein 10 | | Authors: | Ding, J, Wang, D.C. | | Deposit date: | 2010-06-09 | | Release date: | 2010-06-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human programmed cell death 10 complexed with inositol-(1,3,4,5)-tetrakisphosphate: a novel adaptor protein involved in human cerebral cavernous malformation.
Biochem.Biophys.Res.Commun., 399, 2010
|
|
1DUD
 
 | |
1DBP
 
 | |
1VRX
 
 | | Endocellulase e1 from acidothermus cellulolyticus mutant y245g | | Descriptor: | ENDOCELLULASE E1 FROM A. CELLULOLYTICUS | | Authors: | Baker, J.O, McCarley, J.R, Lovett, R, Yu, C.H, Adney, W.S, Rignall, T.R, Vinzant, T.B, Decker, S.R, Sakon, J, Himmel, M.E. | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Catalytically enhanced endocellulase Cel5A from Acidothermus cellulolyticus.
Appl.Biochem.Biotechnol., 121-124, 2005
|
|
