1BMV
 
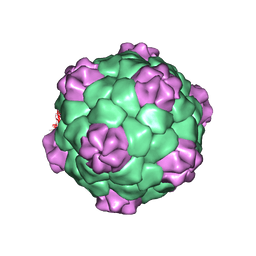 | | PROTEIN-RNA INTERACTIONS IN AN ICOSAHEDRAL VIRUS AT 3.0 ANGSTROMS RESOLUTION | | Descriptor: | PROTEIN (ICOSAHEDRAL VIRUS - A DOMAIN), PROTEIN (ICOSAHEDRAL VIRUS - B AND C DOMAIN), RNA (5'-R(*GP*GP*UP*CP*AP*AP*AP*AP*UP*GP*C)-3') | | Authors: | Chen, Z, Stauffacher, C, Li, Y, Schmidt, T, Bomu, W, Kamer, G, Shanks, M, Lomonossoff, G, Johnson, J.E. | | Deposit date: | 1989-10-09 | | Release date: | 1989-10-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Protein-RNA interactions in an icosahedral virus at 3.0 A resolution.
Science, 245, 1989
|
|
1BMW
 
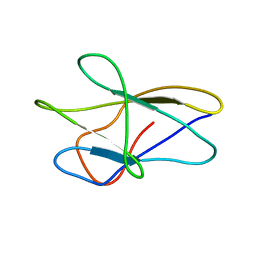 | | A fibronectin type III fold in plant allergens: The solution structure of Phl PII from timothy grass pollen, NMR, 38 STRUCTURES | | Descriptor: | POLLEN ALLERGEN PHL P2 | | Authors: | De Marino, S, Morelli, M.A.C, Fraternali, F, Tamborino, E, Vrtala, S, Dolecek, C, Arosio, P, Valenta, R, Pastore, A. | | Deposit date: | 1998-07-27 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An Immunoglobulin-Like Fold in a Major Plant Allergen: The Solution Structure of Phl P 2 from Timothy Grass Pollen.
Structure Fold.Des., 7, 1999
|
|
1BMX
 
 | |
1BMZ
 
 | |
1BN0
 
 | | SL3 HAIRPIN FROM THE PACKAGING SIGNAL OF HIV-1, NMR, 11 STRUCTURES | | Descriptor: | SL3 RNA HAIRPIN | | Authors: | Pappalardo, L, Kerwood, D.J, Pelczer, I, Borer, P.N. | | Deposit date: | 1998-07-31 | | Release date: | 1999-04-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional folding of an RNA hairpin required for packaging HIV-1.
J.Mol.Biol., 282, 1998
|
|
1BN1
 
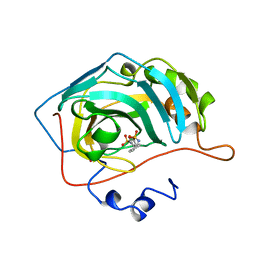 | | CARBONIC ANHYDRASE II INHIBITOR | | Descriptor: | CARBONIC ANHYDRASE, MERCURY (II) ION, THIOPHENE-2,5-DISULFONIC ACID 2-AMIDE-5-(4-METHYL-BENZYLAMIDE), ... | | Authors: | Boriack-Sjodin, P.A, Zeitlin, S, Christianson, D.W. | | Deposit date: | 1998-07-31 | | Release date: | 1999-05-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of inhibitor binding to human carbonic anhydrase II.
Protein Sci., 7, 1998
|
|
1BN3
 
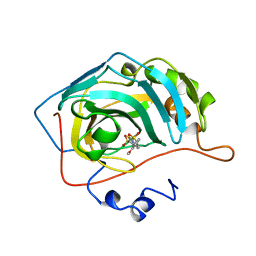 | | CARBONIC ANHYDRASE II INHIBITOR | | Descriptor: | 2-(3-METHOXYPHENYL)-2H-THIENO-[3,2-E]-1,2-THIAZINE-6-SULFINAMIDE-1,1-DIOXIDE, CARBONIC ANHYDRASE, MERCURY (II) ION, ... | | Authors: | Boriack-Sjodin, P.A, Zeitlin, S, Christianson, D.W. | | Deposit date: | 1998-07-31 | | Release date: | 1999-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of inhibitor binding to human carbonic anhydrase II.
Protein Sci., 7, 1998
|
|
1BN4
 
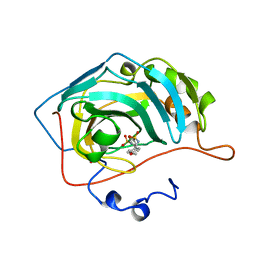 | | CARBONIC ANHYDRASE II INHIBITOR | | Descriptor: | CARBONIC ANHYDRASE, MERCURY (II) ION, N-[(4-METHOXYPHENYL)METHYL]2,5-THIOPHENEDESULFONAMIDE, ... | | Authors: | Boriack-Sjodin, P.A, Zeitlin, S, Christianson, D.W. | | Deposit date: | 1998-07-31 | | Release date: | 1999-05-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of inhibitor binding to human carbonic anhydrase II.
Protein Sci., 7, 1998
|
|
1BN5
 
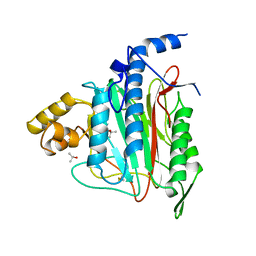 | | HUMAN METHIONINE AMINOPEPTIDASE 2 | | Descriptor: | COBALT (II) ION, METHIONINE AMINOPEPTIDASE, TERTIARY-BUTYL ALCOHOL | | Authors: | Liu, S, Widom, J, Kemp, C.W, Crews, C.M, Clardy, J.C. | | Deposit date: | 1998-07-31 | | Release date: | 1999-07-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human methionine aminopeptidase-2 complexed with fumagillin.
Science, 282, 1998
|
|
1BN6
 
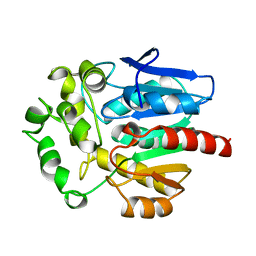 | | HALOALKANE DEHALOGENASE FROM A RHODOCOCCUS SPECIES | | Descriptor: | HALOALKANE DEHALOGENASE | | Authors: | Newman, J, Peat, T.S, Richard, R, Kan, L, Swanson, P.E, Affholter, J.A, Holmes, I.H, Schindler, J.F, Unkefer, C.J, Terwilliger, T.C. | | Deposit date: | 1998-07-31 | | Release date: | 2000-02-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Haloalkane dehalogenases: structure of a Rhodococcus enzyme.
Biochemistry, 38, 1999
|
|
1BN7
 
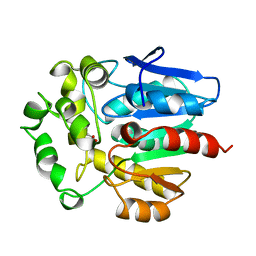 | | HALOALKANE DEHALOGENASE FROM A RHODOCOCCUS SPECIES | | Descriptor: | ACETATE ION, HALOALKANE DEHALOGENASE | | Authors: | Newman, J, Peat, T.S, Richard, R, Kan, L, Swanson, P.E, Affholter, J.A, Holmes, I.H, Schindler, J.F, Unkefer, C.J, Terwilliger, T.C. | | Deposit date: | 1998-07-31 | | Release date: | 2000-02-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Haloalkane dehalogenases: structure of a Rhodococcus enzyme.
Biochemistry, 38, 1999
|
|
1BN8
 
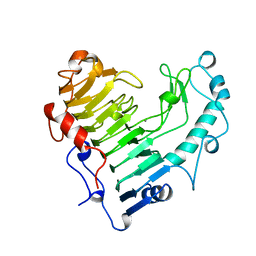 | | BACILLUS SUBTILIS PECTATE LYASE | | Descriptor: | CALCIUM ION, PROTEIN (PECTATE LYASE) | | Authors: | Pickersgill, R, Harris, G, Jenkins, J. | | Deposit date: | 1998-07-31 | | Release date: | 1998-08-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of Bacillus subtilis pectate lyase in complex with calcium.
Nat.Struct.Biol., 1, 1994
|
|
1BN9
 
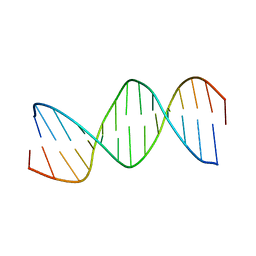 | | RESPONSE ELEMENT OF THE ORPHAN NUCLEAR RECEPTOR REV-ERB BETA | | Descriptor: | DNA (5'-D(*CP*TP*GP*AP*CP*CP*TP*AP*CP*AP*TP*TP*CP*TP*A)-3'), DNA (5'-D(*TP*AP*GP*AP*AP*TP*GP*TP*AP*GP*GP*TP*CP*AP*G)-3') | | Authors: | Castagne, C, Terenzi, H, Zakin, M.M, Delepierre, M. | | Deposit date: | 1998-07-31 | | Release date: | 1998-08-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the orphan nuclear receptor rev-erb beta response element by 1H, 31P NMR and molecular simulation
Biochimie, 82, 2000
|
|
1BNA
 
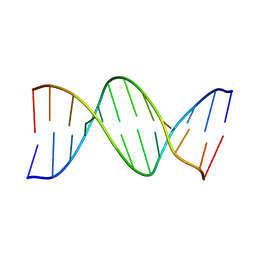 | | STRUCTURE OF A B-DNA DODECAMER. CONFORMATION AND DYNAMICS | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Drew, H.R, Wing, R.M, Takano, T, Broka, C, Tanaka, S, Itakura, K, Dickerson, R.E. | | Deposit date: | 1981-01-26 | | Release date: | 1981-05-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a B-DNA dodecamer: conformation and dynamics.
Proc.Natl.Acad.Sci.USA, 78, 1981
|
|
1BNB
 
 | |
1BNC
 
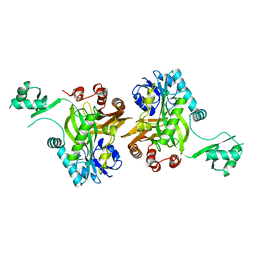 | |
1BND
 
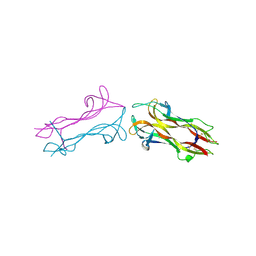 | | STRUCTURE OF THE BRAIN-DERIVED NEUROTROPHIC FACTOR(SLASH)NEUROTROPHIN 3 HETERODIMER | | Descriptor: | BRAIN DERIVED NEUROTROPHIC FACTOR, ISOPROPYL ALCOHOL, NEUROTROPHIN 3 | | Authors: | Robinson, R.C, Radziejewski, C, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1994-12-12 | | Release date: | 1996-04-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the brain-derived neurotrophic factor/neurotrophin 3 heterodimer.
Biochemistry, 34, 1995
|
|
1BNE
 
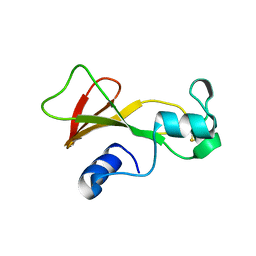 | | BARNASE A43C/S80C DISULFIDE MUTANT | | Descriptor: | BARNASE | | Authors: | Clarke, J, Henrick, K, Fersht, A.R. | | Deposit date: | 1995-03-31 | | Release date: | 1995-07-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Disulfide mutants of barnase. I: Changes in stability and structure assessed by biophysical methods and X-ray crystallography.
J.Mol.Biol., 253, 1995
|
|
1BNF
 
 | | BARNASE T70C/S92C DISULFIDE MUTANT | | Descriptor: | BARNASE | | Authors: | Clarke, J, Henrick, K, Fersht, A.R. | | Deposit date: | 1995-03-31 | | Release date: | 1995-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Disulfide mutants of barnase. I: Changes in stability and structure assessed by biophysical methods and X-ray crystallography.
J.Mol.Biol., 253, 1995
|
|
1BNG
 
 | | BARNASE S85C/H102C DISULFIDE MUTANT | | Descriptor: | BARNASE | | Authors: | Clarke, J, Henrick, K, Fersht, A.R. | | Deposit date: | 1995-03-31 | | Release date: | 1995-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Disulfide mutants of barnase. I: Changes in stability and structure assessed by biophysical methods and X-ray crystallography.
J.Mol.Biol., 253, 1995
|
|
1BNI
 
 | | BARNASE WILDTYPE STRUCTURE AT PH 6.0 | | Descriptor: | BARNASE | | Authors: | Cameron, A, Henrick, K, Fersht, A.R, Dodson, G, Buckle, A.M. | | Deposit date: | 1995-05-17 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structural analysis of mutations in the hydrophobic cores of barnase.
J.Mol.Biol., 234, 1993
|
|
1BNJ
 
 | | BARNASE WILDTYPE STRUCTURE AT PH 9.0 | | Descriptor: | BARNASE | | Authors: | Cameron, A, Henrick, K, Fersht, A.R, Dodson, G, Buckle, A.M. | | Deposit date: | 1995-05-17 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structural analysis of mutations in the hydrophobic cores of barnase.
J.Mol.Biol., 234, 1993
|
|
1BNK
 
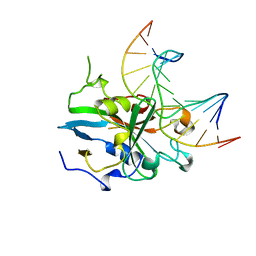 | | HUMAN 3-METHYLADENINE DNA GLYCOSYLASE COMPLEXED TO DNA | | Descriptor: | DNA (5'-D(*GP*AP*CP*AP*TP*GP*YRRP*TP*TP*GP*CP*CP*T)-3'), DNA (5'-D(*GP*GP*CP*AP*AP*TP*CP*AP*TP*GP*TP*CP*A)-3'), PROTEIN (3-METHYLADENINE DNA GLYCOSYLASE) | | Authors: | Lau, A.Y, Schaerer, O.D, Samson, L, Verdine, G.L, Ellenberger, T. | | Deposit date: | 1998-07-29 | | Release date: | 1998-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a human alkylbase-DNA repair enzyme complexed to DNA: mechanisms for nucleotide flipping and base excision.
Cell(Cambridge,Mass.), 95, 1998
|
|
1BNL
 
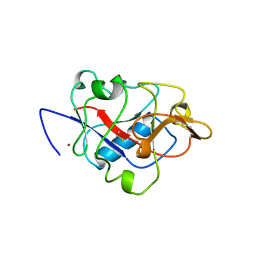 | | ZINC DEPENDENT DIMERS OBSERVED IN CRYSTALS OF HUMAN ENDOSTATIN | | Descriptor: | COLLAGEN XVIII, ZINC ION | | Authors: | Ding, Y.-H, Javaherian, K, Lo, K.-M, Chopra, R, Boehm, T, Lanciotti, J, Harris, B.A, Li, Y, Shapiro, R, Hohenester, E, Timpl, R, Folkman, J, Wiley, D.C. | | Deposit date: | 1998-07-30 | | Release date: | 1998-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Zinc-dependent dimers observed in crystals of human endostatin.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BNM
 
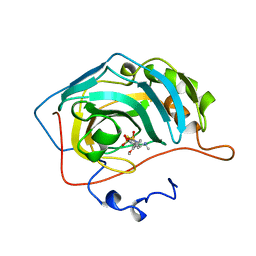 | | CARBONIC ANHYDRASE II INHIBITOR | | Descriptor: | (R)-3,4-DIHYDRO-2-(3-METHOXYPHENYL)-4-METHYLAMINO-2H-THIENO[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE-1,1-DIOXIDE, CARBONIC ANHYDRASE, MERCURY (II) ION, ... | | Authors: | Boriack-Sjodin, P.A, Zeitlin, S, Christianson, D.W. | | Deposit date: | 1998-07-30 | | Release date: | 1999-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of inhibitor binding to human carbonic anhydrase II.
Protein Sci., 7, 1998
|
|
