1ELB
 
 | | Analogous inhibitors of elastase do not always bind analogously | | Descriptor: | 6-ammonio-N-(trifluoroacetyl)-L-norleucyl-N-[4-(1-methylethyl)phenyl]-L-leucinamide, CALCIUM ION, ELASTASE, ... | | Authors: | Mattos, C, Rasmussen, B, Ding, X, Petsko, G.A, Ringe, D. | | Deposit date: | 1993-12-07 | | Release date: | 1994-06-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Analogous inhibitors of elastase do not always bind analogously.
Nat.Struct.Biol., 1, 1994
|
|
1BRG
 
 | |
1FRA
 
 | |
1HLB
 
 | |
1FCT
 
 | |
1XIC
 
 | |
1TYS
 
 | |
1RBL
 
 | | STRUCTURE DETERMINATION AND REFINEMENT OF RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE FROM SYNECHOCOCCUS PCC6301 | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Newman, J, Gutteridge, S, Branden, C.-I, Jones, T.A. | | Deposit date: | 1993-05-12 | | Release date: | 1994-06-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure determination and refinement of ribulose 1,5-bisphosphate carboxylase/oxygenase from Synechococcus PCC6301.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1AHC
 
 | |
1AHA
 
 | | THE N-GLYCOSIDASE MECHANISM OF RIBOSOME-INACTIVATING PROTEINS IMPLIED BY CRYSTAL STRUCTURES OF ALPHA-MOMORCHARIN | | Descriptor: | ADENINE, ALPHA-MOMORCHARIN | | Authors: | Ren, J, Wang, Y, Dong, Y, Stuart, D.I. | | Deposit date: | 1994-01-07 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The N-glycosidase mechanism of ribosome-inactivating proteins implied by crystal structures of alpha-momorcharin.
Structure, 2, 1994
|
|
1AHB
 
 | | THE N-GLYCOSIDASE MECHANISM OF RIBOSOME-INACTIVATING PROTEINS IMPLIED BY CRYSTAL STRUCTURES OF ALPHA-MOMORCHARIN | | Descriptor: | ALPHA-MOMORCHARIN, FORMYCIN-5'-MONOPHOSPHATE | | Authors: | Ren, J, Wang, Y, Dong, Y, Stuart, D.I. | | Deposit date: | 1994-01-07 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The N-glycosidase mechanism of ribosome-inactivating proteins implied by crystal structures of alpha-momorcharin.
Structure, 2, 1994
|
|
1YSC
 
 | |
1GDC
 
 | | REFINED SOLUTION STRUCTURE OF THE GLUCOCORTICOID RECEPTOR DNA-BINDING DOMAIN | | Descriptor: | GLUCOCORTICOID RECEPTOR, ZINC ION | | Authors: | Baumann, H, Paulsen, K, Kovacs, H, Berglund, H, Wright, A.P.H, Gustafsson, J.-A, Hard, T. | | Deposit date: | 1994-03-15 | | Release date: | 1994-06-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of the glucocorticoid receptor DNA-binding domain.
Biochemistry, 32, 1993
|
|
1KFD
 
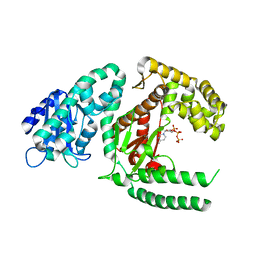 | |
1XIE
 
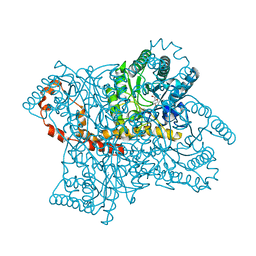 | |
1DLH
 
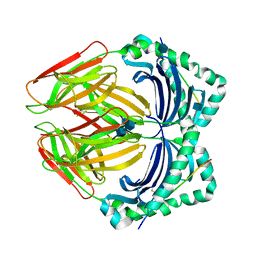 | |
1DGC
 
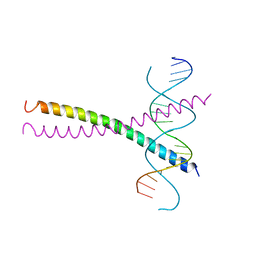 | |
1GLM
 
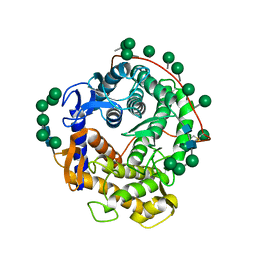 | | REFINED CRYSTAL STRUCTURES OF GLUCOAMYLASE FROM ASPERGILLUS AWAMORI VAR. X100 | | Descriptor: | GLUCOAMYLASE-471, alpha-D-mannopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Aleshin, A.E, Hoffman, C, Firsov, L.M, Honzatko, R.B. | | Deposit date: | 1994-04-25 | | Release date: | 1994-06-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Refined crystal structures of glucoamylase from Aspergillus awamori var. X100.
J.Mol.Biol., 238, 1994
|
|
1GLP
 
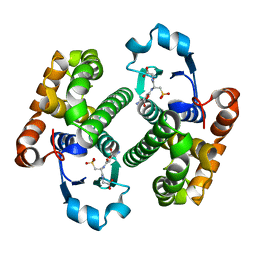 | |
1BPD
 
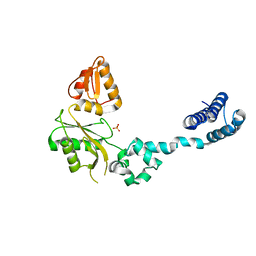 | | CRYSTAL STRUCTURE OF RAT DNA POLYMERASE BETA: EVIDENCE FOR A COMMON POLYMERASE MECHANISM | | Descriptor: | DNA POLYMERASE BETA, PHOSPHATE ION | | Authors: | Sawaya, M.R, Pelletier, H, Kumar, A, Wilson, S.H, Kraut, J. | | Deposit date: | 1994-04-12 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structure of rat DNA polymerase beta: evidence for a common polymerase mechanism.
Science, 264, 1994
|
|
1BPB
 
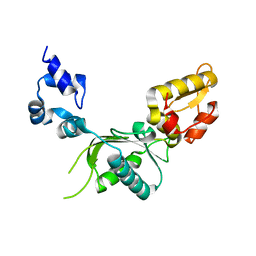 | | CRYSTAL STRUCTURE OF RAT DNA POLYMERASE BETA: EVIDENCE FOR A COMMON POLYMERASE MECHANISM | | Descriptor: | DNA POLYMERASE BETA | | Authors: | Sawaya, M.R, Pelletier, H, Kumar, A, Wilson, S.H, Kraut, J. | | Deposit date: | 1994-04-12 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of rat DNA polymerase beta: evidence for a common polymerase mechanism.
Science, 264, 1994
|
|
1AAQ
 
 | |
1ERA
 
 | |
1GCD
 
 | |
2GDA
 
 | | REFINED SOLUTION STRUCTURE OF THE GLUCOCORTICOID RECEPTOR DNA-BINDING DOMAIN | | Descriptor: | GLUCOCORTICOID RECEPTOR, ZINC ION | | Authors: | Baumann, H, Paulsen, K, Kovacs, H, Berglund, H, Wright, A.P.H, Gustafsson, J.-A, Hard, T. | | Deposit date: | 1994-03-15 | | Release date: | 1994-06-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of the glucocorticoid receptor DNA-binding domain.
Biochemistry, 32, 1993
|
|
