7CG4
 
 | | Cryo-EM structure of the flagellar export apparatus with FliE from Salmonella | | Descriptor: | Flagellar biosynthetic protein FliP, Flagellar hook-basal body complex protein FliE | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2020-06-30 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
2IWF
 
 | | Resting form of pink nitrous oxide reductase from Achromobacter Cycloclastes | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Paraskevopoulos, K, Antonyuk, S.V, Sawers, R.G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2006-06-29 | | Release date: | 2006-08-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Insight Into Catalysis of Nitrous Oxide Reductase from High-Resolution Structures of Resting and Inhibitor-Bound Enzyme from Achromobacter Cycloclastes.
J.Mol.Biol., 362, 2006
|
|
2J0G
 
 | | L-ficolin complexed to N-acetyl-mannosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-mannopyranose, CALCIUM ION, FICOLIN-2, ... | | Authors: | Garlatti, V, Gaboriaud, C. | | Deposit date: | 2006-08-03 | | Release date: | 2007-01-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Insights Into the Innate Immune Recognition Specificities of L- and H-Ficolins.
Embo J., 26, 2007
|
|
2J28
 
 | | MODEL OF E. COLI SRP BOUND TO 70S RNCS | | Descriptor: | 23S RIBOSOMAL RNA, 4.5S SIGNAL RECOGNITION PARTICLE RNA, 50S ribosomal protein L11, ... | | Authors: | Halic, M, Blau, M, Becker, T, Mielke, T, Pool, M.R, Wild, K, Sinning, I, Beckmann, R. | | Deposit date: | 2006-08-16 | | Release date: | 2006-11-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Following the Signal Sequence from Ribosomal Tunnel Exit to Signal Recognition Particle
Nature, 444, 2006
|
|
2IDK
 
 | | Crystal Structure of Rat Glycine N-Methyltransferase Complexed With Folate | | Descriptor: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, Glycine N-methyltransferase | | Authors: | Luka, Z, Pakhomova, S, Loukachevitch, L.V, Egli, M, Newcomer, M.E, Wagner, C. | | Deposit date: | 2006-09-15 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | 5-methyltetrahydrofolate is bound in intersubunit areas of rat liver folate-binding protein glycine N-methyltransferase.
J.Biol.Chem., 282, 2007
|
|
2IDV
 
 | | Crystal structure of wheat C113S mutant EIF4E bound TO 7-methyl-GDP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Eukaryotic translation initiation factor 4E-1 | | Authors: | Monzingo, A.F, Dutt-Chaudhuri, A, Sadow, J, Dhaliwal, S, Hoffman, D.W, Robertus, J.D, Browning, K.S. | | Deposit date: | 2006-09-15 | | Release date: | 2007-06-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of eukaryotic translation initiation factor-4E from wheat reveals a novel disulfide bond.
Plant Physiol., 143, 2007
|
|
7COU
 
 | | Structure of cyanobacterial photosystem II in the dark S1 state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Shen, J.-R, Suga, M. | | Deposit date: | 2020-08-05 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Capturing structural changes of the S 1 to S 2 transition of photosystem II using time-resolved serial femtosecond crystallography.
Iucrj, 8, 2021
|
|
7CJG
 
 | | Structural and kinetic characterization of Porphyromonas gingivalis glutaminyl cyclase | | Descriptor: | 5,6-DIMETHYLBENZIMIDAZOLE, GLYCEROL, Glutamine cyclotransferase-related protein, ... | | Authors: | Ruiz-Carrillo, D, Lamers, S, Feng, Q, Yu, S, Sun, B, Jiang, J, Lukman, M. | | Deposit date: | 2020-07-10 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and kinetic characterization of Porphyromonas gingivalis glutaminyl cyclase.
Biol.Chem., 402, 2021
|
|
2IHP
 
 | | Yeast inorganic pyrophosphatase with magnesium and phosphate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Inorganic pyrophosphatase, MAGNESIUM ION, ... | | Authors: | Oksanen, E, Ahonen, A.K, Tuominen, H, Tuominen, V, Lahti, R, Goldman, A, Heikinheimo, P. | | Deposit date: | 2006-09-27 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Complete Structural Description of the Catalytic Cycle of Yeast Pyrophosphatase.
Biochemistry, 46, 2007
|
|
7CJE
 
 | | Structural and kinetic characterization of Porphyromonas gingivalis glutaminyl cyclase | | Descriptor: | GLYCEROL, Glutamine cyclotransferase-related protein, MAGNESIUM ION, ... | | Authors: | Ruiz-Carrillo, D, Lamers, S, Feng, Q, Yu, S, Sun, B, Jiang, J, Lukman, M. | | Deposit date: | 2020-07-10 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.950007 Å) | | Cite: | Structural and kinetic characterization of Porphyromonas gingivalis glutaminyl cyclase.
Biol.Chem., 402, 2021
|
|
7C2X
 
 | | Crystal Structure of Glycyrrhiza uralensis UGT73P12 complexed with glycyrrhetinic acid 3-O-monoglucuronide | | Descriptor: | (2~{S},3~{S},4~{S},5~{R},6~{R})-6-[[(3~{S},4~{a}~{R},6~{a}~{R},6~{b}~{S},8~{a}~{S},11~{S},12~{a}~{R},14~{a}~{R},14~{b}~{S})-11-carboxy-4,4,6~{a},6~{b},8~{a},11,14~{b}-heptamethyl-14-oxidanylidene-2,3,4~{a},5,6,7,8,9,10,12,12~{a},14~{a}-dodecahydro-1~{H}-picen-3-yl]oxy]-3,4,5-tris(oxidanyl)oxane-2-carboxylic acid, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Ren, J. | | Deposit date: | 2020-05-09 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of Glycyrrhiza uralensis UGT73P12 complexed with glycyrrhetinic acid 3-O-monoglucuronide
To Be Published
|
|
2IK0
 
 | | Yeast inorganic pyrophosphatase variant E48D with magnesium and phosphate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Inorganic pyrophosphatase, MAGNESIUM ION, ... | | Authors: | Oksanen, E, Ahonen, A.K, Tuominen, H, Tuominen, V, Lahti, R, Goldman, A, Heikinheimo, P. | | Deposit date: | 2006-10-02 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Complete Structural Description of the Catalytic Cycle of Yeast Pyrophosphatase.
Biochemistry, 46, 2007
|
|
7C3O
 
 | | Crystal structure of TT109 from CANDIDA ALBICANS | | Descriptor: | ACETYL COENZYME *A, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Chen, Y.P, Lei, J.H, Lu, D.R, Su, D. | | Deposit date: | 2020-05-13 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of TT109 from CANDIDA ALBICANS
To Be Published
|
|
7CH7
 
 | | Cryo-EM structure of E.coli MlaFEB | | Descriptor: | Lipid asymmetry maintenance ABC transporter permease subunit MlaE, Lipid asymmetry maintenance protein MlaB, Phospholipid ABC transporter ATP-binding protein MlaF | | Authors: | Zhou, C, Shi, H, Huang, Y. | | Deposit date: | 2020-07-05 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Insight into Phospholipid Transport by the MlaFEBD Complex from P. aeruginosa.
J.Mol.Biol., 433, 2021
|
|
2IUW
 
 | | Crystal structure of human ABH3 in complex with iron ion and 2- oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, ALKYLATED REPAIR PROTEIN ALKB HOMOLOG 3, BETA-MERCAPTOETHANOL, ... | | Authors: | Sundheim, O, Vagbo, C.B, Bjoras, M, deSousa, M.M.L, Talstad, V, Aas, P.A, Drablos, F, Krokan, H.E, Tainer, J.A, Slupphaug, G. | | Deposit date: | 2006-06-07 | | Release date: | 2006-07-26 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Human Abh3 Structure and Key Residues for Oxidative Demethylation to Reverse DNA/RNA Damage.
Embo J., 25, 2006
|
|
2IK4
 
 | | Yeast inorganic pyrophosphatase variant D117E with magnesium and phosphate | | Descriptor: | Inorganic pyrophosphatase, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Oksanen, E, Ahonen, A.K, Tuominen, H, Tuominen, V, Lahti, R, Goldman, A, Heikinheimo, P. | | Deposit date: | 2006-10-02 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Complete Structural Description of the Catalytic Cycle of Yeast Pyrophosphatase.
Biochemistry, 46, 2007
|
|
2IME
 
 | | 2-Hydroxychromene-2-carboxylate Isomerase: a Kappa Class Glutathione-S-Transferase from Pseudomonas putida | | Descriptor: | (2S)-2-HYDROXY-2H-CHROMENE-2-CARBOXYLIC ACID, (3E)-4-(2-HYDROXYPHENYL)-2-OXOBUT-3-ENOIC ACID, 2-hydroxychromene-2-carboxylate isomerase, ... | | Authors: | Thompson, L.C, Ladner, J.E, Codreanu, S.G, Harp, J, Gilliland, G.L, Armstrong, R.N. | | Deposit date: | 2006-10-04 | | Release date: | 2007-06-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 2-Hydroxychromene-2-carboxylic Acid Isomerase: A Kappa Class Glutathione Transferase from Pseudomonas putida
Biochemistry, 46, 2007
|
|
2IZ8
 
 | | MS2-RNA HAIRPIN (C-7) COMPLEX | | Descriptor: | MS2 COAT PROTEIN, RNA, (5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*CP*UP*CP* AP*CP*CP*CP*AP*UP*GP*U)-3') | | Authors: | Helgstrand, C, Grahn, E, Stonehouse, N.J, Stockley, P.G, Liljas, L. | | Deposit date: | 2006-07-25 | | Release date: | 2006-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Investigating the Structural Basis of Purine Specificity in the Structures of MS2 Coat Protein RNA Translational Operator Hairpins
Nucleic Acids Res., 30, 2002
|
|
2IDH
 
 | |
2FUD
 
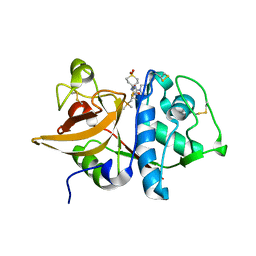 | | Human Cathepsin S with Inhibitor CRA-27566 | | Descriptor: | N-{(1R)-2-[(4-CYANO-1,1-DIOXIDOTETRAHYDRO-2H-THIOPYRAN-4-YL)AMINO]-2-OXO-1-[(TRIMETHYLSILYL)METHYL]ETHYL}MORPHOLINE-4-CARBOXAMIDE, cathepsin S | | Authors: | Somoza, J.R. | | Deposit date: | 2006-01-26 | | Release date: | 2006-08-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Human Cathepsin S with Inhibitor CRA-27566
To be Published
|
|
2FRQ
 
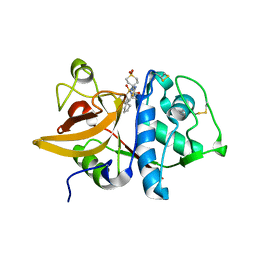 | | Human Cathepsin S with Inhibitor CRA-26871 | | Descriptor: | N-[4-(AMINOMETHYL)-1,1-DIOXIDOTETRAHYDRO-2H-THIOPYRAN-4-YL]-3-(1-METHYLCYCLOPENTYL)-N~2~-[(1E)-N-(PHENYLSULFONYL)ETHANIMIDOYL]-L-ALANINAMIDE, cathepsin S | | Authors: | Somoza, J.R. | | Deposit date: | 2006-01-19 | | Release date: | 2006-07-25 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Human Cathepsin S with Inhibitor CRA-26871
To be Published
|
|
2G6L
 
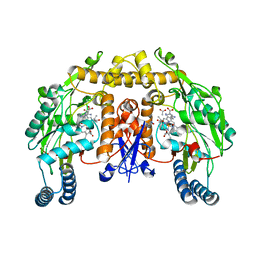 | | Structure of rat nNOS heme domain (BH2 bound) complexed with NO | | Descriptor: | 7,8-DIHYDROBIOPTERIN, ACETATE ION, ARGININE, ... | | Authors: | Li, H, Igarashi, J, Jamal, J, Yang, W, Poulos, T.L. | | Deposit date: | 2006-02-24 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural studies of constitutive nitric oxide synthases with diatomic ligands bound.
J.Biol.Inorg.Chem., 11, 2006
|
|
2G74
 
 | |
2ID7
 
 | | 1.75 A Structure of T87I Phosphono-CheY | | Descriptor: | Chemotaxis protein cheY | | Authors: | Halkides, C.J, Haas, R.M, McAdams, K.A, Casper, E.S, Santarsiero, B.D, Mesecar, A.D. | | Deposit date: | 2006-09-14 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structures of T87I phosphono-CheY and T87I/Y106W phosphono-CheY help to explain their binding affinities to the FliM and CheZ peptides.
Arch.Biochem.Biophys., 479, 2008
|
|
2IK6
 
 | | Yeast inorganic pyrophosphatase variant D120E with magnesium and phosphate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Inorganic pyrophosphatase, MAGNESIUM ION, ... | | Authors: | Oksanen, E, Ahonen, A.K, Tuominen, H, Tuominen, V, Lahti, R, Goldman, A, Heikinheimo, P. | | Deposit date: | 2006-10-02 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Complete Structural Description of the Catalytic Cycle of Yeast Pyrophosphatase.
Biochemistry, 46, 2007
|
|
