8VQY
 
 | | Human GABAA receptor alpha1-beta2-gamma2 subtype in complex with GABA plus methaqualone | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-methyl-3-(2-methylphenyl)quinazolin-4(3H)-one, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Chojnacka, W, Teng, J, Kim, J.J, Jensen, A.A, Hibbs, R.E. | | Deposit date: | 2024-01-19 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Structural insights into GABA A receptor potentiation by Quaalude.
Nat Commun, 15, 2024
|
|
8VE9
 
 | | IsPETase - ACCCETN mutant - CombiPETase | | Descriptor: | CHLORIDE ION, GLYCEROL, Poly(ethylene terephthalate) hydrolase | | Authors: | Joho, Y, Royan, S, Newton, S, Caputo, A.T, Ardevol Grau, A, Jackson, C. | | Deposit date: | 2023-12-18 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enhancing PET Degrading Enzymes: A Combinatory Approach.
Chembiochem, 25, 2024
|
|
7CRO
 
 | | NSD2 bearing E1099K/T1150A dual mutation in complex with 187-bp NCP | | Descriptor: | DNA (168-MER), Histone H2A, Histone H2B, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
8VLU
 
 | | Cryo-EM structure of human HGSNAT bound with CoA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COENZYME A, Heparan-alpha-glucosaminide N-acetyltransferase | | Authors: | Li, F, Zhao, B. | | Deposit date: | 2024-01-12 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structural and mechanistic insights into a lysosomal membrane enzyme HGSNAT involved in Sanfilippo syndrome
To Be Published
|
|
2FW4
 
 | | Carbonic anhydrase activators. The first X-ray crystallographic study of an activator of isoform I, structure with L-histidine. | | Descriptor: | Carbonic anhydrase 1, HISTIDINE, ZINC ION | | Authors: | Temperini, C, Scozzafava, A, Supuran, C.T. | | Deposit date: | 2006-02-01 | | Release date: | 2006-08-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Carbonic anhydrase activators: The first X-ray crystallographic study of an adduct of isoform I.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
8VLY
 
 | | Cryo-EM structure of human HGSNAT in transition state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heparan-alpha-glucosaminide N-acetyltransferase | | Authors: | Li, F, Zhao, B. | | Deposit date: | 2024-01-12 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Structural and mechanistic insights into a lysosomal membrane enzyme HGSNAT involved in Sanfilippo syndrome
To Be Published
|
|
2FYX
 
 | |
8UGP
 
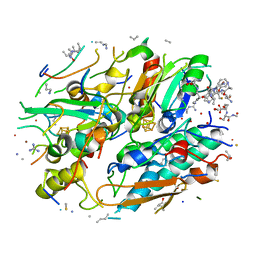 | | In-situ structure of typeA supercomplex in respiratory chain ( local refined map focused on CI iron-sulfur cluster regions ) | | Descriptor: | IRON/SULFUR CLUSTER, NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 12, NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 6, ... | | Authors: | Zheng, W, Zhang, K, Zhu, J. | | Deposit date: | 2023-10-05 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | High-resolution in situ structures of mammalian respiratory supercomplexes.
Nature, 631, 2024
|
|
8VEM
 
 | | IsPETase - ACCE mutant | | Descriptor: | Poly(ethylene terephthalate) hydrolase, SULFATE ION | | Authors: | Joho, Y, Royan, S, Newton, S, Caputo, A.T, Ardevol Grau, A, Jackson, C. | | Deposit date: | 2023-12-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.711 Å) | | Cite: | Enhancing PET Degrading Enzymes: A Combinatory Approach.
Chembiochem, 25, 2024
|
|
7COY
 
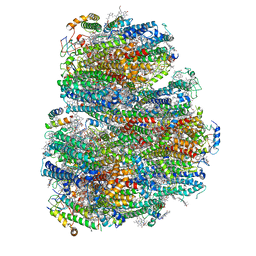 | | Structure of the far-red light utilizing photosystem I of Acaryochloris marina | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, CHLOROPHYLL D, ... | | Authors: | Kawakami, K, Yonekura, K, Hamaguchi, T, Kashino, Y, Shinzawa-Itoh, K, Inoue-Kashino, N, Itoh, S, Ifuku, K, Yamashita, E. | | Deposit date: | 2020-08-05 | | Release date: | 2021-03-31 | | Last modified: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure of the far-red light utilizing photosystem I of Acaryochloris marina.
Nat Commun, 12, 2021
|
|
8UKF
 
 | |
8VLI
 
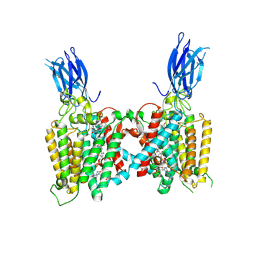 | | Cryo-EM structure of human HGSNAT bound with CoA and product analog | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-methyl-2-oxo-2H-1-benzopyran-7-yl 2-acetamido-2-deoxy-beta-D-glucopyranoside, ... | | Authors: | Li, F, Zhao, B. | | Deposit date: | 2024-01-11 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and mechanistic insights into a lysosomal membrane enzyme HGSNAT involved in Sanfilippo syndrome
To Be Published
|
|
2FMM
 
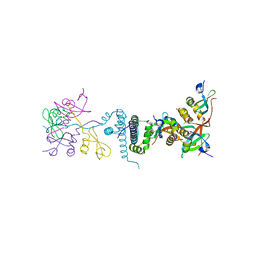 | | Crystal Structure of EMSY-HP1 complex | | Descriptor: | Chromobox protein homolog 1, Protein EMSY, SULFATE ION | | Authors: | Huang, Y. | | Deposit date: | 2006-01-09 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the HP1-EMSY complex reveals an unusual mode of HP1 binding.
Structure, 14, 2006
|
|
7CB0
 
 | |
7CB5
 
 | |
7COH
 
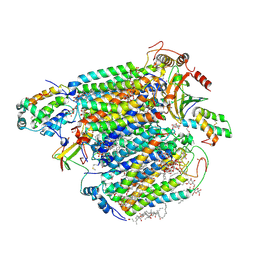 | | Dimeric Form of Bovine Heart Cytochrome c Oxidase in the Fully Oxidized State | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, 1,2-ETHANEDIOL, ... | | Authors: | Shinzawa-Itoh, K, Muramoto, K. | | Deposit date: | 2020-08-04 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The 1.3-A Resolution structure of bovine cytochrome c oxidase suggests a dimerization mechanism
Biochim.Biophys.Acta, 2021
|
|
8UKD
 
 | |
8VEL
 
 | | IsPETase - ACCCC mutant | | Descriptor: | Poly(ethylene terephthalate) hydrolase, SULFATE ION | | Authors: | Joho, Y, Royan, S, Newton, S, Caputo, A.T, Ardevol Grau, A, Jackson, C. | | Deposit date: | 2023-12-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.624 Å) | | Cite: | Enhancing PET Degrading Enzymes: A Combinatory Approach.
Chembiochem, 25, 2024
|
|
2IDR
 
 | | Crystal structure of translation initiation factor EIF4E from wheat | | Descriptor: | Eukaryotic translation initiation factor 4E-1 | | Authors: | Monzingo, A.F, Sadow, J, Dhaliwal, S, Lyon, A, Hoffman, D.W, Robertus, J.D, Browning, K.S. | | Deposit date: | 2006-09-15 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of eukaryotic translation initiation factor-4E from wheat reveals a novel disulfide bond.
Plant Physiol., 143, 2007
|
|
7CB6
 
 | |
2INE
 
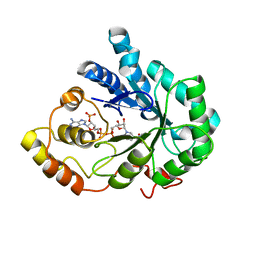 | |
2IKG
 
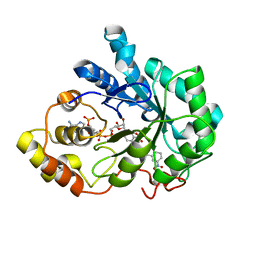 | | Aldose reductase complexed with nitrophenyl-oxadiazol type inhibitor at 1.43 A | | Descriptor: | 4-[3-(3-NITROPHENYL)-1,2,4-OXADIAZOL-5-YL]BUTANOIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Steuber, H, Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2006-10-02 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural and Thermodynamic Study on Aldose Reductase: Nitro-substituted Inhibitors with Strong Enthalpic Binding Contribution
J.Mol.Biol., 368, 2007
|
|
7CGS
 
 | |
2IS7
 
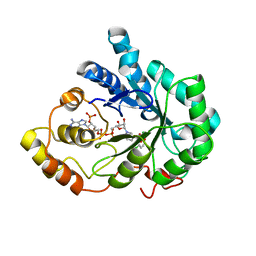 | | Crystal Structure of Aldose Reductase complexed with Dichlorophenylacetic acid | | Descriptor: | (2,6-DICHLOROPHENYL)ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H.T, Milne, A, Brownlee, J.M. | | Deposit date: | 2006-10-16 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and thermodynamic studies of simple aldose reductase-inhibitor complexes.
Bioorg.Chem., 34, 2006
|
|
7CGO
 
 | | Cryo-EM structure of the flagellar motor-hook complex from Salmonella | | Descriptor: | Flagellar L-ring protein, Flagellar M-ring protein, Flagellar MS ring L1, ... | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2020-07-01 | | Release date: | 2021-04-28 | | Last modified: | 2021-05-26 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
