9FX6
 
 | | Crystal structure of Cryo2RT SARS-CoV-2 main protease at 100K | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Huang, C.Y, Aumonier, S, Mac Sweeney, A, Olieric, V, Wang, M. | | Deposit date: | 2024-07-01 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.227 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8VG7
 
 | | Crystal Structure of T115A Variant of D-Dopachrome Tautomerase (D-DT) | | Descriptor: | D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Wolff, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-23 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
8XVC
 
 | | CryoEM structure of ADP-DNA-MuB conformation1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent target DNA activator B | | Authors: | Zhao, X, Zhang, K, Li, S. | | Deposit date: | 2024-01-14 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Elucidating the Architectural dynamics of MuB filaments in bacteriophage Mu DNA transposition.
Nat Commun, 15, 2024
|
|
8VEQ
 
 | | Crystal structure of transpeptidase domain of PBP2 from Neisseria gonorrhoeae cephalosporin-resistant strain H041 in complex with azlocillin | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-2-oxo-1-{[(2R)-2-{[(2-oxoimidazolidin-1-yl)carbonyl]amino}-2-phenylacetyl]amino}ethyl]-1,3-thiazolidine-4-carboxylic acid, Probable peptidoglycan D,D-transpeptidase PenA | | Authors: | Stratton, C, Bala, S, Davies, C. | | Deposit date: | 2023-12-20 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ureidopenicillins Are Potent Inhibitors of Penicillin-Binding Protein 2 from Multidrug-Resistant Neisseria gonorrhoeae H041.
Acs Infect Dis., 10, 2024
|
|
9AXG
 
 | |
9EYR
 
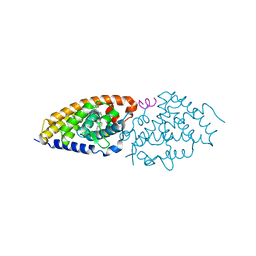 | | VDR complex with gemini analog UG-480 | | Descriptor: | (1~{R},3~{S},5~{Z})-5-[(2~{E})-2-[(1~{R},3~{a}~{S},7~{a}~{R})-7~{a}-methyl-1-[(1~{R})-5-methyl-1-[(1~{S},2~{S})-2-(3-methyl-3-oxidanyl-butyl)cyclopropyl]-5-oxidanyl-hexyl]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N. | | Deposit date: | 2024-04-09 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of New Type of Gemini Analogues with a Cyclopropane Moiety in Their Side Chain.
J.Med.Chem., 67, 2024
|
|
8YKU
 
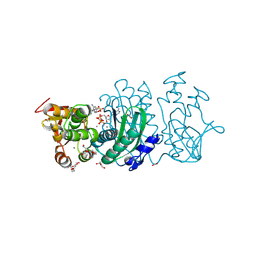 | |
8VRK
 
 | | Rigid body fitted model for refined density map of gamma tubulin ring complex capped microtubule | | Descriptor: | Actin, cytoplasmic 1, Gamma-tubulin complex component 3, ... | | Authors: | Aher, A, Urnavicius, L, Kapoor, T.M. | | Deposit date: | 2024-01-22 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Structure of the gamma-tubulin ring complex-capped microtubule.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8XT3
 
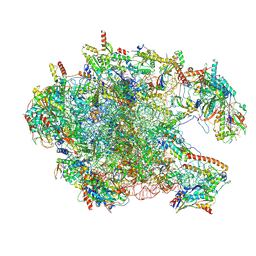 | | Cryo-EM structure of the human 39S mitoribosome with 10uM Tigecycline | | Descriptor: | 16s rRNA, 39S ribosomal protein L22, mitochondrial, ... | | Authors: | Li, X, Wang, M, Cheng, J. | | Deposit date: | 2024-01-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
9EM2
 
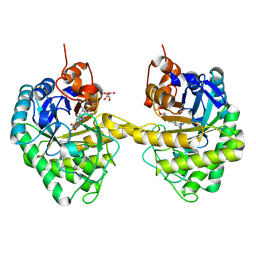 | |
8WBS
 
 | | Crystal structure of cis-Epoxysuccinate Hydrolases KlCESH[L]-D48N complexed with sulfate ions | | Descriptor: | (S)-2-haloacid dehalogenase, CALCIUM ION, SULFATE ION | | Authors: | Dong, S, Xuan, J.S, Feng, Y.G, Cui, Q. | | Deposit date: | 2023-09-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid.
J.Biol.Chem., 300, 2024
|
|
8XIA
 
 | |
8WU4
 
 | | Cryo-EM structure of native H. thermoluteolus TH-1 GroEL | | Descriptor: | Chaperonin GroEL | | Authors: | Liao, Z, Gopalasingam, C.C, Kameya, M, Gerle, C, Shigematsu, H, Ishii, M, Arakawa, T, Fushinobu, S. | | Deposit date: | 2023-10-20 | | Release date: | 2024-03-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into thermophilic chaperonin complexes.
Structure, 32, 2024
|
|
8Y7M
 
 | | FluPol-NS2 complex (local refinement) | | Descriptor: | Nuclear export protein, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Peng, Q, Sun, J.Q. | | Deposit date: | 2024-02-04 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | NS2 stabilizes polymerase hexamer to regulate transcription-replication switch of influenza A virus
To Be Published
|
|
8VBU
 
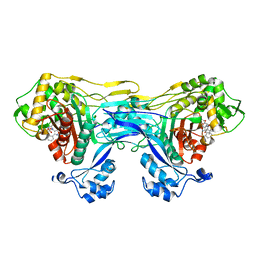 | | Structure of the monofunctional Staphylococcus aureus PBP1 in its beta-lactam (Oxacillin) inhibited form | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-{[(5-methyl-3-phenyl-1,2-oxazol-4-yl)carbonyl]amino}-2-oxoethyl]-1,3-thiazolidine-4-carb oxylic acid, Penicillin-binding protein 1 | | Authors: | Bon, C.G, Lee, J, Caveney, N.A, Strynadka, N.C.J. | | Deposit date: | 2023-12-12 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and kinetic analysis of the monofunctional Staphylococcus aureus PBP1.
J.Struct.Biol., 216, 2024
|
|
8VLX
 
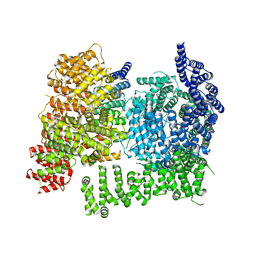 | | HTT in complex with HAP40 and a small molecule. | | Descriptor: | 40-kDa huntingtin-associated protein, Huntingtin, N-[(1S,2R)-2-benzylcyclopentyl]-N'-{1-[(1S)-1-(pyridin-4-yl)ethyl]piperidin-4-yl}urea | | Authors: | Poweleit, N, Boudet, J, Doherty, E. | | Deposit date: | 2024-01-12 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Discovery of a Small Molecule Ligand to the Huntingtin/HAP40 complex
To Be Published
|
|
9B8D
 
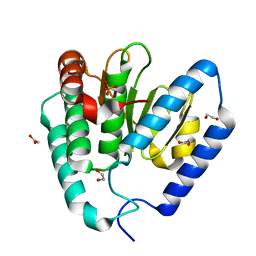 | | Structure of Legionella pneumophila Ceg10 | | Descriptor: | 1,2-ETHANEDIOL, Ceg10, PHOSPHATE ION | | Authors: | Tomchick, D.R, Heisler, D.B, Alto, N.M. | | Deposit date: | 2024-03-29 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Exploiting bacterial effector proteins to uncover evolutionarily conserved antiviral host machinery.
Plos Pathog., 20, 2024
|
|
8YFK
 
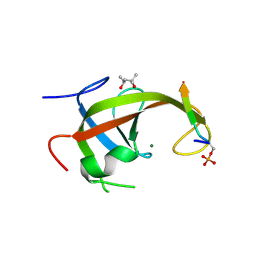 | | Crystal structure of FIP200 claw/TNIP1_FIR_pS123 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MAGNESIUM ION, RB1-inducible coiled-coil protein 1, ... | | Authors: | Lv, M.Q, Wu, S.M. | | Deposit date: | 2024-02-24 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for TNIP1 binding to FIP200 during mitophagy.
J.Biol.Chem., 2024
|
|
8VH7
 
 | | Crystal structure of heparosan synthase 2 from Pasteurella multocida at 1.98 A | | Descriptor: | 1,2-ETHANEDIOL, Heparosan synthase B, MANGANESE (II) ION, ... | | Authors: | Pedersen, L.C, Liu, J, Stancanelli, E, Krahn, J.M. | | Deposit date: | 2023-12-31 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | Structural and Functional Analysis of Heparosan Synthase 2 from Pasteurella multocida to Improve the Synthesis of Heparin
Acs Catalysis, 14, 2024
|
|
8VHM
 
 | | Structure of DHODH in Complex with Fragment 2 | | Descriptor: | ACETATE ION, CHLORIDE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2024-01-02 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Discovery of Alternative Binding Poses through Fragment-Based Identification of DHODH Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
8VG5
 
 | | Crystal Structure of V113N Variant of D-Dopachrome Tautomerase (D-DT) Bound with 4CPPC | | Descriptor: | 4-(3-carboxyphenyl)pyridine-2,5-dicarboxylic acid, CITRIC ACID, D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-22 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
8ZH4
 
 | | HIV-1 integrase core domain in complex with compound 5 | | Descriptor: | (2~{S})-2-(4',5-dimethylspiro[1,2-dihydroindene-3,1'-cyclohexane]-4-yl)-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, Integrase, SULFATE ION, ... | | Authors: | Furuzono, T, Orita, T, Nomura, A, Adachi, T. | | Deposit date: | 2024-05-10 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Design and synthesis of novel and potent allosteric HIV-1 integrase inhibitors with a spirocyclic moiety.
Bioorg.Med.Chem.Lett., 110, 2024
|
|
9EX0
 
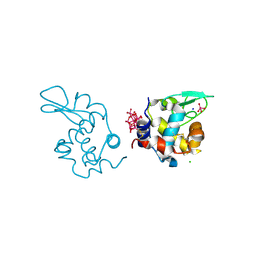 | | X-ray structure of a polyoxidovanadate/lysozyme adduct obtained when the protein is treated with [VIVO(acac)2] in 1.1 M NaCl, 0.1 M sodium acetate at pH 4.0 (Structure A) | | Descriptor: | CHLORIDE ION, Lysozyme C, Polyoxidovanadate complex, ... | | Authors: | Tito, G, Merlino, A, Ferraro, G. | | Deposit date: | 2024-04-05 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Non-Covalent and Covalent Binding of New Mixed-Valence Cage-like Polyoxidovanadate Clusters to Lysozyme.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8WBO
 
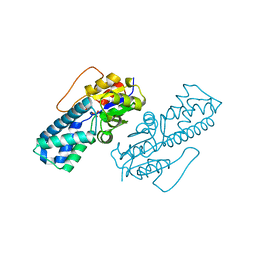 | | Crystal structure of cis-Epoxysuccinate Hydrolases RhCESH[L] mutant D18N complexed with sulfate ions | | Descriptor: | Epoxide hydrolase, SULFATE ION | | Authors: | Dong, S, Xuan, J.S, Feng, Y.G, Cui, Q. | | Deposit date: | 2023-09-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid.
J.Biol.Chem., 300, 2024
|
|
8XN8
 
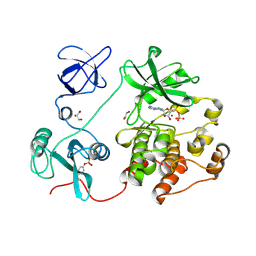 | | The Crystal Structure of SRC from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Bao, C. | | Deposit date: | 2023-12-29 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of SRC from Biortus.
To Be Published
|
|
