8CCJ
 
 | |
8CCM
 
 | |
8CCL
 
 | |
5HN0
 
 | | Dengue serotype 3 RNA-dependent RNA polymerase bound to compound 4 | | Descriptor: | (6-hydroxybiphenyl-3-yl)acetic acid, RNA-directed RNA polymerase NS5, ZINC ION | | Authors: | Noble, C.G. | | Deposit date: | 2016-01-17 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Potent Non-Nucleoside Inhibitors of Dengue Viral RNA-Dependent RNA Polymerase from a Fragment Hit Using Structure-Based Drug Design.
J.Med.Chem., 59, 2016
|
|
4UIP
 
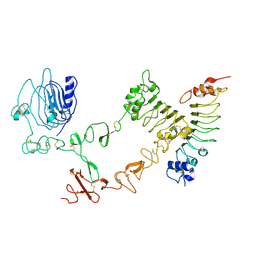 | | The complex structure of extracellular domain of EGFR with Repebody (rAC1). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, EPIDERMAL GROWTH FACTOR RECEPTOR, ... | | Authors: | Kang, Y.J, Cha, Y.J, Cho, H.S, Lee, J.J, Kim, H.S. | | Deposit date: | 2015-03-31 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Enzymatic Prenylation and Oxime Ligation for the Synthesis of Stable and Homogeneous Protein-Drug Conjugates for Targeted Therapy.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4UOW
 
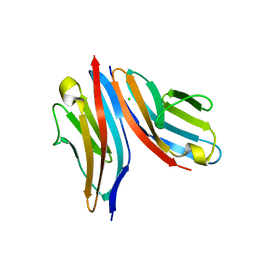 | | Crystal structure of the titin M10-Obscurin Ig domain 1 complex | | Descriptor: | CHLORIDE ION, Obscurin, SODIUM ION, ... | | Authors: | Pernigo, S, Fukuzawa, A, Gautel, M, Steiner, R.A. | | Deposit date: | 2014-06-10 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Crystal Structure of the Human Titin:Obscurin Complex Reveals a Conserved Yet Specific Muscle M-Band Zipper Module.
J.Mol.Biol., 427, 2015
|
|
5IIT
 
 | |
6RG0
 
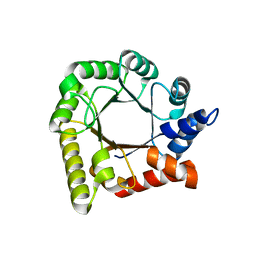 | | Structure of pdxj | | Descriptor: | Pyridoxine 5'-phosphate synthase | | Authors: | Rohweder, B, Rajendran, C, Sterner, R. | | Deposit date: | 2019-04-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.074 Å) | | Cite: | Library Selection with a Randomized Repertoire of ( beta alpha )8-Barrel Enzymes Results in Unexpected Induction of Gene Expression.
Biochemistry, 58, 2019
|
|
6S4B
 
 | | Crystal Structure of BRD4(1) bound to inhibitor BUX1 (8) | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Bromodomain-containing protein 4, CALCIUM ION, ... | | Authors: | Huegle, M. | | Deposit date: | 2019-06-27 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 4-Acyl Pyrroles as Dual BET-BRD7/9 Bromodomain Inhibitors Address BETi Insensitive Human Cancer Cell Lines.
J.Med.Chem., 63, 2020
|
|
6S6K
 
 | | Crystal Structure of BRD4(1) bound to inhibitor BUX2 (9) | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Bromodomain-containing protein 4, ~{N}-[5-(azepan-1-ylsulfonyl)-2-methoxy-phenyl]-3-methyl-4-oxidanylidene-5,6,7,8-tetrahydro-2~{H}-cyclohepta[c]pyrrole-1-carboxamide | | Authors: | Huegle, M. | | Deposit date: | 2019-07-03 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 4-Acyl Pyrroles as Dual BET-BRD7/9 Bromodomain Inhibitors Address BETi Insensitive Human Cancer Cell Lines.
J.Med.Chem., 63, 2020
|
|
5IO3
 
 | | Crystal structure of the legionella pneumophila effector protein RavZ - I422 | | Descriptor: | Uncharacterized protein RavZ | | Authors: | Kwon, D.H, Kim, L, Kim, B.-W, Hong, S.B, Song, H.K. | | Deposit date: | 2016-03-08 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | The 1:2 complex between RavZ and LC3 reveals a mechanism for deconjugation of LC3 on the phagophore membrane
Autophagy, 13, 2017
|
|
5IJH
 
 | |
5IIQ
 
 | |
5IJJ
 
 | |
5IJP
 
 | |
6SA2
 
 | | Crystal Structure of BRD4(1) bound to inhibitor BUX3 (10) | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-methoxy-5-morpholin-4-ylsulfonyl-phenyl)-3-methyl-4-oxidanylidene-5,6,7,8-tetrahydro-2~{H}-cyclohepta[c]pyrrole-1-carboxamide | | Authors: | Huegle, M. | | Deposit date: | 2019-07-16 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 4-Acyl Pyrroles as Dual BET-BRD7/9 Bromodomain Inhibitors Address BETi Insensitive Human Cancer Cell Lines.
J.Med.Chem., 63, 2020
|
|
6S8Z
 
 | |
6RWJ
 
 | | Crystal Structure of BRD4(1) bound to inhibitor BUG0 (6) | | Descriptor: | Bromodomain-containing protein 4, ~{N},3-dimethyl-4-oxidanylidene-5,6,7,8-tetrahydro-2~{H}-cyclohepta[c]pyrrole-1-carboxamide | | Authors: | Huegle, M. | | Deposit date: | 2019-06-05 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 4-Acyl Pyrroles as Dual BET-BRD7/9 Bromodomain Inhibitors Address BETi Insensitive Human Cancer Cell Lines.
J.Med.Chem., 63, 2020
|
|
6SAH
 
 | | Crystal Structure of BRD4(1) bound to inhibitor BUX5 (11) | | Descriptor: | Bromodomain-containing protein 4, ~{N}-(2-methoxy-5-piperidin-1-ylsulfonyl-phenyl)-3-methyl-4-oxidanylidene-5,6,7,8-tetrahydro-2~{H}-cyclohepta[c]pyrrole-1-carboxamide | | Authors: | Huegle, M. | | Deposit date: | 2019-07-16 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 4-Acyl Pyrroles as Dual BET-BRD7/9 Bromodomain Inhibitors Address BETi Insensitive Human Cancer Cell Lines.
J.Med.Chem., 63, 2020
|
|
6SA3
 
 | | Crystal Structure of BRD4(1) bound to inhibitor BUX4 (13) | | Descriptor: | Bromodomain-containing protein 4, ~{N}-[2-methoxy-5-(4-methylpiperazin-1-yl)sulfonyl-phenyl]-3-methyl-4-oxidanylidene-5,6,7,8-tetrahydro-2~{H}-cyclohepta[c]pyrrole-1-carboxamide | | Authors: | Huegle, M. | | Deposit date: | 2019-07-16 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 4-Acyl Pyrroles as Dual BET-BRD7/9 Bromodomain Inhibitors Address BETi Insensitive Human Cancer Cell Lines.
J.Med.Chem., 63, 2020
|
|
5J7N
 
 | | Crystal structure of a small heat-shock protein from Xylella fastidiosa reveals a distinct high order structure | | Descriptor: | Low molecular weight heat shock protein | | Authors: | Fonseca, E.M.B, Scorsato, V, dos Santos, C.A, Tomazini Jr, A, Aparicio, R, Polikarpov, I. | | Deposit date: | 2016-04-06 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a small heat-shock protein from Xylella fastidiosa reveals a distinct high-order structure.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
6SB8
 
 | | Crystal Structure of BRD4(1) bound to inhibitor BUX14 (7) | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Bromodomain-containing protein 4, ~{N}-[5-(diethylsulfamoyl)-2-oxidanyl-phenyl]-3-methyl-4-oxidanylidene-5,6,7,8-tetrahydro-2~{H}-cyclohepta[c]pyrrole-1-carboxamide | | Authors: | Huegle, M. | | Deposit date: | 2019-07-19 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 4-Acyl Pyrroles as Dual BET-BRD7/9 Bromodomain Inhibitors Address BETi Insensitive Human Cancer Cell Lines.
J.Med.Chem., 63, 2020
|
|
5HX8
 
 | |
5I76
 
 | | Crystal structure of FM318, a recombinant Fab adopted from cetuximab | | Descriptor: | FM318_heavy_cahin, FM318_light_chain | | Authors: | Sim, D.W, Kim, J.H, Seok, S.H, Seo, M.D, Kim, Y.P, Won, H.S. | | Deposit date: | 2016-02-16 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Bacterial production and structure-functional validation of a recombinant antigen-binding fragment (Fab) of an anti-cancer therapeutic antibody targeting epidermal growth factor receptor.
Appl.Microbiol.Biotechnol., 100, 2016
|
|
6SAJ
 
 | | Crystal Structure of BRD4(1) bound to inhibitor BUX6 (12) | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Bromodomain-containing protein 4, ~{N}-[2-methoxy-5-(2-oxa-6-azaspiro[3.3]heptan-6-ylsulfonyl)phenyl]-3-methyl-4-oxidanylidene-5,6,7,8-tetrahydro-2~{H}-cyclohepta[c]pyrrole-1-carboxamide | | Authors: | Huegle, M. | | Deposit date: | 2019-07-16 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 4-Acyl Pyrroles as Dual BET-BRD7/9 Bromodomain Inhibitors Address BETi Insensitive Human Cancer Cell Lines.
J.Med.Chem., 63, 2020
|
|
