4G85
 
 | | Crystal structure of human HisRS | | Descriptor: | Histidine-tRNA ligase, cytoplasmic | | Authors: | Wei, Z, Wu, J, Zhou, J.J, Yang, X.-L, Zhang, M, Schimmel, P. | | Deposit date: | 2012-07-21 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Internally Deleted Human tRNA Synthetase Suggests Evolutionary Pressure for Repurposing.
Structure, 20, 2012
|
|
5DMB
 
 | |
5DMG
 
 | | X-RAY STRUCTURE OF THE FAB FRAGMENT OF THE ANTI TAU ANTIBODY RB86 IN COMPLEX WITH THE PHOSPHORYLATED TAU PEPTIDE (416-430) | | Descriptor: | Microtubule-associated protein, RB86 antibody Fab fragment heavy chain, RB86 antibody Fab fragment light chain | | Authors: | Benz, J, Lorenz, S, Georges, G, Jochner, A, Goepfert, U, Grueninger, F, Bujotzek, A. | | Deposit date: | 2015-09-08 | | Release date: | 2015-12-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | VH-VL orientation prediction for antibody humanization candidate selection: A case study.
Mabs, 8, 2016
|
|
5DOB
 
 | | Crystal structure of the Human Cytomegalovirus Nuclear Egress Complex (NEC) | | Descriptor: | CALCIUM ION, Virion egress protein UL31 homolog, Virion egress protein UL34 homolog, ... | | Authors: | Lye, M.F, El Omari, K, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-09-11 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Unexpected features and mechanism of heterodimer formation of a herpesvirus nuclear egress complex.
Embo J., 34, 2015
|
|
4G8L
 
 | | Active state of intact sensor domain of human RNase L with 2-5A bound | | Descriptor: | 2-5A-dependent ribonuclease, 5'-O-MONOPHOSPHORYLADENYLYL(2'->5')ADENYLYL(2'->5')ADENOSINE | | Authors: | Han, Y, Whitney, G, Donovan, J, Korennykh, A. | | Deposit date: | 2012-07-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Innate Immune Messenger 2-5A Tethers Human RNase L into Active High-Order Complexes.
Cell Rep, 2, 2012
|
|
4GA1
 
 | |
6TIR
 
 | |
2BTY
 
 | | Acetylglutamate kinase from Thermotoga maritima complexed with its inhibitor arginine | | Descriptor: | ACETYLGLUTAMATE KINASE, ARGININE, N-ACETYL-L-GLUTAMATE, ... | | Authors: | Gil-Ortiz, F, Fernandez-Murga, M.L, Fita, I, Rubio, V. | | Deposit date: | 2005-06-08 | | Release date: | 2005-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Bases of Feed-Back Control of Arginine Biosynthesis, Revealed by the Structure of Two Hexameric N-Acetylglutamate Kinases, from Thermotoga Maritima and Pseudomonas Aeruginosa
J.Mol.Biol., 356, 2006
|
|
8P66
 
 | |
5E1P
 
 | | Ca(2+)-Calmodulin from Paramecium tetraurelia qFit disorder model | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Calmodulin | | Authors: | Lin, J, van den Bedem, H, Brunger, A.T, Wilson, M.A. | | Deposit date: | 2015-09-29 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Atomic resolution experimental phase information reveals extensive disorder and bound 2-methyl-2,4-pentanediol in Ca(2+)-calmodulin.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
2BWR
 
 | | Crystal Structure of Psathyrella Velutina Lectin at 1.5A Resolution | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Cioci, G, Mitchell, E.P, Chazalet, V, Gautier, C, Oscarson, S, Debray, H, Perez, S, Imberty, A. | | Deposit date: | 2005-07-18 | | Release date: | 2006-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Beta-Propeller Crystal Structure of Psathyrella Velutina Lectin: An Integrin-Like Fungal Protein Interacting with Monosaccharides and Calcium.
J.Mol.Biol., 357, 2006
|
|
4G0H
 
 | | Crystal structure of the N-terminal domain of Helicobacter pylori CagA protein | | Descriptor: | Cytotoxicity-associated immunodominant antigen | | Authors: | Kaplan-Turkoz, B, Remaut, H, Dian, C, Louche, A, Terradot, L. | | Deposit date: | 2012-07-09 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural insights into Helicobacter pylori oncoprotein CagA interaction with beta 1 integrin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4G20
 
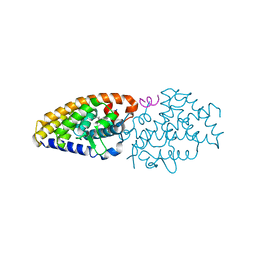 | | Structural basis for the accommodation of bis- and tris-aromatic derivatives in Vitamin D Nuclear Receptor | | Descriptor: | 3-(5'-{2-[3,4-bis(hydroxymethyl)phenyl]ethyl}-2'-methyl-2-propylbiphenyl-4-yl)pentan-3-ol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Ciesielski, F, Sato, Y, Moras, D, Rochel, N. | | Deposit date: | 2012-07-11 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the accommodation of bis- and tris-aromatic derivatives in vitamin d nuclear receptor.
J.Med.Chem., 55, 2012
|
|
2C1F
 
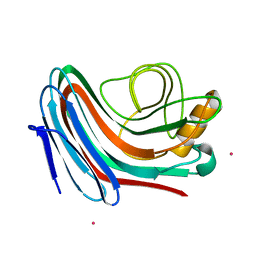 | |
4G38
 
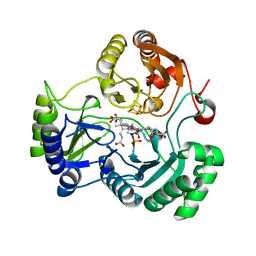 | |
5DM9
 
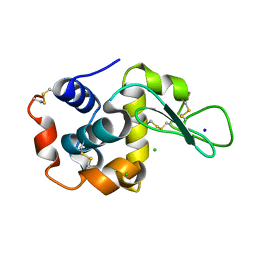 | |
8SW7
 
 | | BG505 Boost2 SOSIP.664 in complex with NHP polyclonal antibody FP1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 Boost 2 gp120, BG505 Boost 2 gp41, ... | | Authors: | Pratap, P.P, Antansijevic, A, Ozorowski, G, Ward, A.B. | | Deposit date: | 2023-05-17 | | Release date: | 2023-07-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Focusing antibody responses to the fusion peptide in rhesus macaques.
Biorxiv, 2023
|
|
5DQT
 
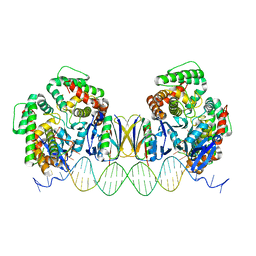 | | Crystal Structure of Cas-DNA-22 complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (33-MER), ... | | Authors: | Wang, J, Li, J, Zhao, H, Sheng, G, Wang, M, Yin, M, Wang, Y. | | Deposit date: | 2015-09-15 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and Mechanistic Basis of PAM-Dependent Spacer Acquisition in CRISPR-Cas Systems.
Cell, 163, 2015
|
|
4GAC
 
 | |
5E8R
 
 | | Human HMT1 hnRNP methyltransferase-like protein 6 (S. cerevisiae) | | Descriptor: | CHLORIDE ION, N-methyl-N-({4-[4-(propan-2-yloxy)phenyl]-1H-pyrrol-3-yl}methyl)ethane-1,2-diamine, Protein arginine N-methyltransferase 6, ... | | Authors: | DONG, A, ZENG, H, LIU, J, TEMPEL, W, Seitova, A, Hutchinson, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, JIN, J, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-10-14 | | Release date: | 2015-12-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A Potent, Selective, and Cell-Active Inhibitor of Human Type I Protein Arginine Methyltransferases.
Acs Chem.Biol., 11, 2016
|
|
5E93
 
 | | Crystal Structure of Trypanosoma cruzi Dihydroorotate Dehydrogenase in Complex with Neq0071 | | Descriptor: | 1,2-ETHANEDIOL, 5-[(E)-3-(furan-2-yl)prop-2-enylidene]-1,3-diazinane-2,4,6-trione, CACODYLATE ION, ... | | Authors: | Rocha, J.R, Inaoka, D.K, Cheleski, J, Shiba, T, Harada, S, Montanari, C.A, Kita, K. | | Deposit date: | 2015-10-14 | | Release date: | 2016-10-19 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Exploring Trypanosoma cruzi Dihydroorotate Dehydrogenase Active Site Plasticity for the Discovery of Potent and Selective Inhibitors with Trypanocidal Activity
To be Published
|
|
5DX0
 
 | |
5DXJ
 
 | | Crystal structure of CARM1 and sinefungin | | Descriptor: | GLYCEROL, Histone-arginine methyltransferase CARM1, SINEFUNGIN | | Authors: | Boriack-Sjodin, P.A. | | Deposit date: | 2015-09-23 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into Ternary Complex Formation of Human CARM1 with Various Substrates.
Acs Chem.Biol., 11, 2016
|
|
4FYB
 
 | | Structural and functional characterizations of a thioredoxin-fold protein from Helicobacter pylori | | Descriptor: | GLYCEROL, Thiol:disulfide interchange protein (DsbC) | | Authors: | Yoon, J.Y, Kim, J, Lee, S.J, Im, H.N, Kim, H.S, Yoon, H, An, D.R, Kim, J.Y, Kim, S, Han, B.W, Suh, S.W. | | Deposit date: | 2012-07-04 | | Release date: | 2013-05-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional characterization of HP0377, a thioredoxin-fold protein from Helicobacter pylori
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4FZ7
 
 | |
