1RFP
 
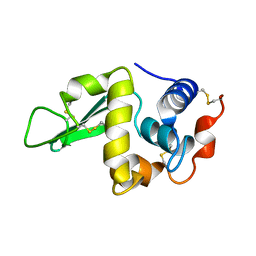 | |
1RFQ
 
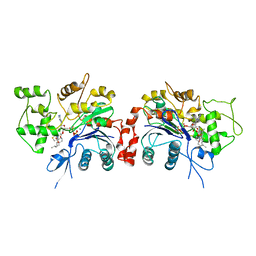 | | Actin Crystal Dynamics: Structural Implications for F-actin Nucleation, Polymerization and Branching Mediated by the Anti-parallel Dimer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Reutzel, R, Yoshioka, C, Govindasamy, L, Yarmola, E.G, Agbandje-McKenna, M, Bubb, M.R, McKenna, R. | | Deposit date: | 2003-11-10 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Actin crystal dynamics: structural implications for F-actin nucleation, polymerization, and branching mediated by the anti-parallel dimer.
J.Struct.Biol., 146, 2004
|
|
1RFR
 
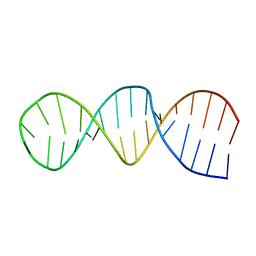 | | NMR structure of the 30mer stemloop-D of coxsackieviral RNA | | Descriptor: | stemloop-D RNA of the 5'-cloverleaf of coxsackievirus B3 | | Authors: | Ohlenschlager, O, Wohnert, J, Bucci, E, Seitz, S, Hafner, S, Ramachandran, R, Zell, R, Gorlach, M. | | Deposit date: | 2003-11-10 | | Release date: | 2004-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the stemloop D subdomain of coxsackievirus B3 cloverleaf
RNA and its interaction with the proteinase 3C.
STRUCTURE, 12, 2004
|
|
1RFS
 
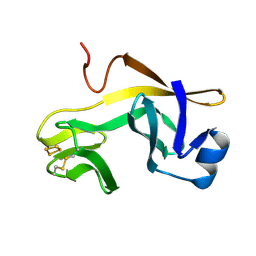 | | RIESKE SOLUBLE FRAGMENT FROM SPINACH | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, RIESKE PROTEIN | | Authors: | Carrell, C.J, Zhang, H, Cramer, W.A, Smith, J.L. | | Deposit date: | 1997-08-14 | | Release date: | 1998-01-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Biological identity and diversity in photosynthesis and respiration: structure of the lumen-side domain of the chloroplast Rieske protein.
Structure, 5, 1997
|
|
1RFT
 
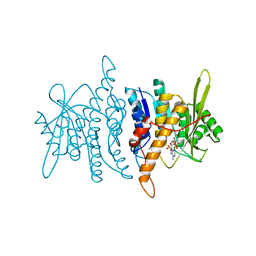 | | Crystal structure of pyridoxal kinase complexed with AMP-PCP and pyridoxamine | | Descriptor: | 4-(AMINOMETHYL)-5-(HYDROXYMETHYL)-2-METHYLPYRIDIN-3-OL, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Liang, D.-C, Jiang, T, Li, M.-H. | | Deposit date: | 2003-11-10 | | Release date: | 2004-04-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational changes in the reaction of pyridoxal kinase
J.BIOL.CHEM., 279, 2004
|
|
1RFU
 
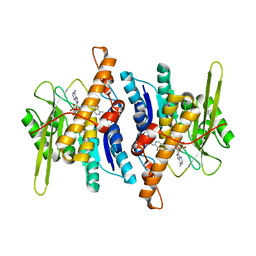 | | Crystal structure of pyridoxal kinase complexed with ADP and PLP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PYRIDOXAL-5'-PHOSPHATE, ZINC ION, ... | | Authors: | Liang, D.-C, Jiang, T, Li, M.-H. | | Deposit date: | 2003-11-10 | | Release date: | 2004-04-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational changes in the reaction of pyridoxal kinase
J.BIOL.CHEM., 279, 2004
|
|
1RFV
 
 | |
1RFX
 
 | | Crystal Structure of resisitin | | Descriptor: | ACETATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Patel, S.D, Rajala, M.W, Scherer, P.E, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-11-10 | | Release date: | 2004-06-08 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Disulfide-dependent multimeric assembly of resistin family hormones
Science, 304, 2004
|
|
1RFY
 
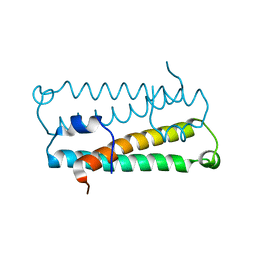 | | Crystal Structure of Quorum-Sensing Antiactivator TraM | | Descriptor: | Transcriptional repressor traM | | Authors: | Chen, G, Malenkos, J.W, Cha, M.R, Fuqua, C, Chen, L. | | Deposit date: | 2003-11-10 | | Release date: | 2004-11-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Quorum-sensing antiactivator TraM forms a dimer that dissociates to inhibit TraR
Mol.Microbiol., 52, 2004
|
|
1RFZ
 
 | | Structure of Protein of Unknown Function from Bacillus stearothermophilus | | Descriptor: | Hypothetical protein APC35681, SULFATE ION | | Authors: | Kim, Y, Wu, R, Cuff, M.E, Quartey, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-11-10 | | Release date: | 2004-03-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of hypothetical protein APC35681 from Bacillus stearothermophilus
To be Published
|
|
1RG0
 
 | |
1RG1
 
 | | Crystal structure of human Tyrosyl-DNA Phosphodiesterase complexed with vanadate, octopamine, and tetranucleotide AGTT | | Descriptor: | 4-(2S-AMINO-1-HYDROXYETHYL)PHENOL, 5'-D(*AP*GP*TP*T)-3', SPERMINE, ... | | Authors: | Davies, D.R, Interthal, H, Champoux, J.J, Hol, W.G. | | Deposit date: | 2003-11-10 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Explorations of peptide and oligonucleotide binding sites of tyrosyl-DNA phosphodiesterase using vanadate complexes.
J.Med.Chem., 47, 2004
|
|
1RG2
 
 | | Crystal structure of human Tyrosyl-DNA Phosphodiesterase complexed with vanadate, octopamine, and tetranucleotide AGTA | | Descriptor: | 4-(2R-AMINO-1-HYDROXYETHYL)PHENOL, 4-(2S-AMINO-1-HYDROXYETHYL)PHENOL, 5'-D(*AP*GP*TP*A)-3', ... | | Authors: | Davies, D.R, Interthal, H, Champoux, J.J, Hol, W.G. | | Deposit date: | 2003-11-11 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Explorations of peptide and oligonucleotide binding sites of tyrosyl-DNA phosphodiesterase using vanadate complexes.
J.Med.Chem., 47, 2004
|
|
1RG3
 
 | |
1RG4
 
 | |
1RG5
 
 | | Structure of the photosynthetic reaction centre from Rhodobacter sphaeroides carotenoidless strain R-26.1 | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Roszak, A.W, Hashimoto, H, Gardiner, A.T, Cogdell, R.J, Isaacs, N.W. | | Deposit date: | 2003-11-11 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protein Regulation of Carotenoid Binding: Gatekeeper and Locking Amino Acid Residues in Reaction Centers of Rhodobacter sphaeroides
STRUCTURE, 12, 2004
|
|
1RG6
 
 | | Solution structure of the C-terminal domain of p63 | | Descriptor: | second splice variant p63 | | Authors: | Cadot, B, Candi, E, Cicero, D.O, Desideri, A, Mele, S, Melino, G, Paci, M. | | Deposit date: | 2003-11-11 | | Release date: | 2004-11-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal domain of p63
To be Published
|
|
1RG7
 
 | | DIHYDROFOLATE REDUCTASE COMPLEXED WITH METHOTREXATE | | Descriptor: | DIHYDROFOLATE REDUCTASE, METHOTREXATE | | Authors: | Sawaya, M.R, Kraut, J. | | Deposit date: | 1996-11-27 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Loop and subdomain movements in the mechanism of Escherichia coli dihydrofolate reductase: crystallographic evidence.
Biochemistry, 36, 1997
|
|
1RG8
 
 | |
1RG9
 
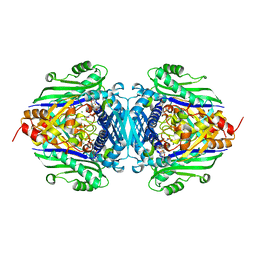 | | S-Adenosylmethionine synthetase complexed with SAM and PPNP | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Komoto, J, Yamada, T, Takata, Y, Markham, G.D, Takusagawa, F. | | Deposit date: | 2003-11-11 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the S-adenosylmethionine synthetase ternary complex: a novel catalytic mechanism of s-adenosylmethionine synthesis from ATP and MET.
Biochemistry, 43, 2004
|
|
1RGA
 
 | |
1RGB
 
 | | Phospholipase A2 from Vipera ammodytes meridionalis | | Descriptor: | (9E)-OCTADEC-9-ENAMIDE, Phospholipase A2 | | Authors: | Georgieva, D.N. | | Deposit date: | 2003-11-12 | | Release date: | 2005-01-18 | | Last modified: | 2018-10-03 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Asp49 phospholipase A(2)-elaidoylamide complex: a new mode of inhibition.
Biochem.Biophys.Res.Commun., 319, 2004
|
|
1RGC
 
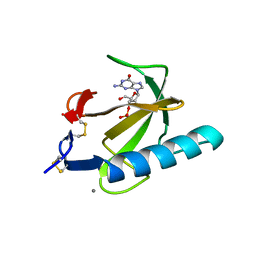 | | THE COMPLEX BETWEEN RIBONUCLEASE T1 AND 3'-GUANYLIC ACID SUGGESTS GEOMETRY OF ENZYMATIC REACTION PATH. AN X-RAY STUDY | | Descriptor: | CALCIUM ION, GUANOSINE-3'-MONOPHOSPHATE, RIBONUCLEASE T1 | | Authors: | Heydenreich, A, Koellner, G, Choe, H.W, Cordes, F, Kisker, C, Schindelin, H, Adamiak, R, Hahn, U, Saenger, W. | | Deposit date: | 1993-05-12 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The complex between ribonuclease T1 and 3'GMP suggests geometry of enzymic reaction path. An X-ray study.
Eur.J.Biochem., 218, 1993
|
|
1RGD
 
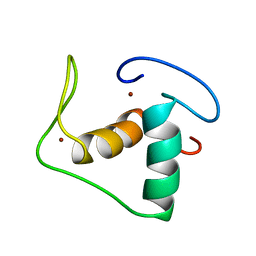 | | STRUCTURE REFINEMENT OF THE GLUCOCORTICOID RECEPTOR-DNA BINDING DOMAIN FROM NMR DATA BY RELAXATION MATRIX CALCULATIONS | | Descriptor: | GLUCOCORTICOID RECEPTOR, ZINC ION | | Authors: | Van Tilborg, M.A.A, Bonvin, A.M.J.J, Hard, K, Davis, A, Maler, B, Boelens, R, Yamamoto, K.R, Kaptein, R. | | Deposit date: | 1995-01-06 | | Release date: | 1995-02-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure refinement of the glucocorticoid receptor-DNA binding domain from NMR data by relaxation matrix calculations.
J.Mol.Biol., 247, 1995
|
|
1RGE
 
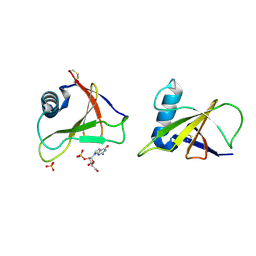 | | HYDROLASE, GUANYLORIBONUCLEASE | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE, SULFATE ION | | Authors: | Sevcik, J, Dauter, Z, Lamzin, V.S, Wilson, K.S. | | Deposit date: | 1995-06-05 | | Release date: | 1996-10-14 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Ribonuclease from Streptomyces aureofaciens at atomic resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
