8FMH
 
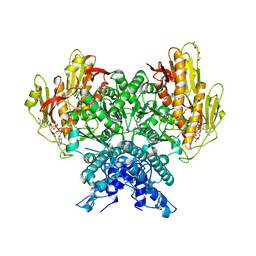 | | Structure of CBASS Cap5 from Pseudomonas syringae as an activated tetramer with the cyclic dinucleotide 3'2'-c-dGAMP ligand (2 tetramers in the AU) | | Descriptor: | 3'2'-cGAMP, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Rechkoblit, O, Kreitler, D.F, Aggarwal, A.K. | | Deposit date: | 2022-12-23 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Activation of CBASS Cap5 endonuclease immune effector by cyclic nucleotides.
Nat.Struct.Mol.Biol., 31, 2024
|
|
1Z79
 
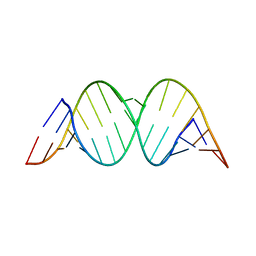 | |
5Y20
 
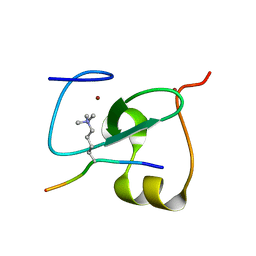 | |
5OOM
 
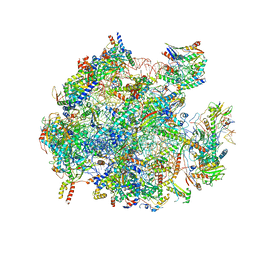 | | Structure of a native assembly intermediate of the human mitochondrial ribosome with unfolded interfacial rRNA | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Brown, A, Rathore, S, Kimanius, D, Aibara, S, Bai, X.C, Rorbach, J, Amunts, A, Ramakrishnan, V. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-13 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structures of the human mitochondrial ribosome in native states of assembly.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5Q1P
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A in complex with FMOPL000299a | | Descriptor: | 1-cyclohexyl-3-(2-pyridin-4-ylethyl)urea, DNA cross-link repair 1A protein, MALONATE ION, ... | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-15 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5Q27
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A in complex with FMOPL000421a | | Descriptor: | 5-ethyl-~{N}-[(1-methylpyrazol-4-yl)methyl]thiophene-2-carboxamide, DIMETHYL SULFOXIDE, DNA cross-link repair 1A protein, ... | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-15 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5Q1W
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A in complex with FMOPL000546a | | Descriptor: | 1~{H}-benzimidazol-2-ylcyanamide, DNA cross-link repair 1A protein, MALONATE ION, ... | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-15 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5Q25
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A in complex with FMOPL000532a | | Descriptor: | 1~{H}-indol-6-yl-(4-methylpiperazin-1-yl)methanone, DNA cross-link repair 1A protein, MALONATE ION, ... | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-15 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5Q24
 
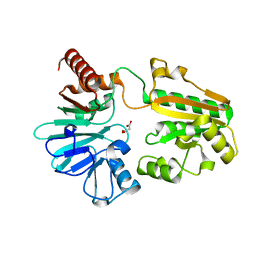 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A in complex with FMOPL000676a | | Descriptor: | 4-[[[(4~{S})-2,2-dimethyloxan-4-yl]amino]methyl]phenol, DNA cross-link repair 1A protein, MALONATE ION, ... | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-15 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5Q1X
 
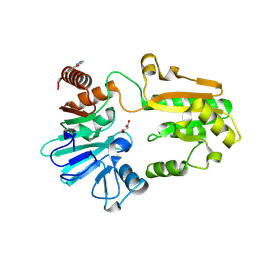 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A in complex with FMOPL000085a | | Descriptor: | 2,4-bis(fluoranyl)-6-(1~{H}-pyrazol-3-yl)phenol, DNA cross-link repair 1A protein, MALONATE ION, ... | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-15 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
6XQJ
 
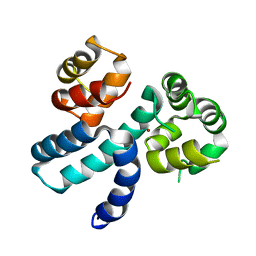 | | Structure of HIV-1 Vpr in complex with the human nucleotide excision repair protein hHR23A | | Descriptor: | Protein Vpr,UV excision repair protein RAD23 homolog A, ZINC ION | | Authors: | Byeon, I.-J.L, Calero, G, Wu, Y, Byeon, C.H, Gronenborn, A.M. | | Deposit date: | 2020-07-09 | | Release date: | 2021-11-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of HIV-1 Vpr in complex with the human nucleotide excision repair protein hHR23A.
Nat Commun, 12, 2021
|
|
5NJ5
 
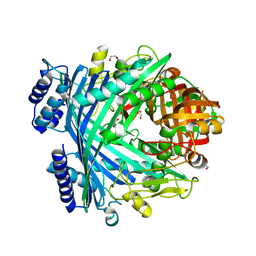 | | E. coli Microcin-processing metalloprotease TldD/E with phosphate bound | | Descriptor: | 1,2-ETHANEDIOL, Metalloprotease PmbA, Metalloprotease TldD, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
6LUF
 
 | |
2KQH
 
 | | A G-rich sequence within the c-kit oncogene promoter forms a parallel G-quadruplex having asymmetric G-tetrad dynamics | | Descriptor: | 5'-D(*CP*GP*GP*GP*CP*GP*GP*GP*CP*GP*CP*GP*AP*GP*GP*GP*AP*GP*GP*GP*T)-3', POTASSIUM ION | | Authors: | Hsu, S.-T.D, Varnai, P, Bugaut, A, Reszka, A.P, Neidle, S, Balasubramanian, S. | | Deposit date: | 2009-11-05 | | Release date: | 2009-11-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A G-rich sequence within the c-kit oncogene promoter forms a parallel G-quadruplex having asymmetric G-tetrad dynamics
J.Am.Chem.Soc., 131, 2009
|
|
3ZQJ
 
 | | Mycobacterium tuberculosis UvrA | | Descriptor: | UVRABC SYSTEM PROTEIN A, ZINC ION | | Authors: | Rossi, F, Khanduja, J.S, Bortoluzzi, A, Houghton, J, Sander, P, Guthlein, C, Davis, E.O, Springer, B, Bottger, E.C, Relini, A, Penco, A, Muniyappa, K, Rizzi, M. | | Deposit date: | 2011-06-09 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Biological and Structural Characterization of Mycobacterium Tuberculosis Uvra Provides Novel Insights Into its Mechanism of Action
Nucleic Acids Res., 39, 2011
|
|
6YES
 
 | |
1YZD
 
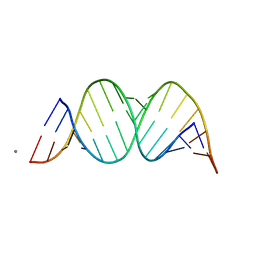 | |
5NJ9
 
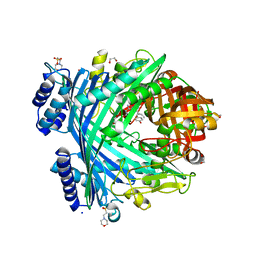 | | E. coli Microcin-processing metalloprotease TldD/E with DRVY angiotensin fragment bound | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ASP-ARG-VAL-TYR, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
6C34
 
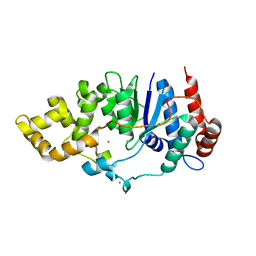 | | Mycobacterium smegmatis DNA flap endonuclease mutant D125N | | Descriptor: | 5'-3' exonuclease, MANGANESE (II) ION | | Authors: | Shuman, S, Goldgur, Y, Carl, A, Uson, M.L. | | Deposit date: | 2018-01-09 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and mutational analysis of Mycobacterium smegmatis FenA highlight active site amino acids and three metal ions essential for flap endonuclease and 5' exonuclease activities.
Nucleic Acids Res., 46, 2018
|
|
1GD1
 
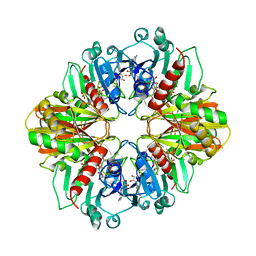 | |
4J5S
 
 | | TARG1 (C6orf130), Terminal ADP-ribose Glycohydrolase 1 ADP-ribose complex | | Descriptor: | 1,2-ETHANEDIOL, 1,3,2-DIOXABOROLAN-2-OL, BORATE ION, ... | | Authors: | Schellenberg, M.J, Appel, C.D, Krahn, J, Williams, R.S. | | Deposit date: | 2013-02-09 | | Release date: | 2013-03-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease.
Embo J., 32, 2013
|
|
8H68
 
 | | Crystal structure of Caenorhabditis elegans NMAD-1 in complex with NOG and Mg(II) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shi, Y, Ding, J, Yang, H. | | Deposit date: | 2022-10-16 | | Release date: | 2023-02-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Caenorhabditis elegans NMAD-1 functions as a demethylase for actin.
J Mol Cell Biol, 15, 2023
|
|
5Q1R
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A in complex with FMOPL000291a | | Descriptor: | 3-(azepan-1-ylmethyl)-1~{H}-indole, DNA cross-link repair 1A protein, MALONATE ION, ... | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-15 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5Q20
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A in complex with FMOPL000524a | | Descriptor: | 1-[2-methyl-1,3-bis(oxidanyl)propan-2-yl]-3-phenyl-urea, DNA cross-link repair 1A protein, MALONATE ION, ... | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-15 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
6B43
 
 | | CryoEM structure and atomic model of the Kaposi's sarcoma-associated herpesvirus capsid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Dai, X.H, Gong, D.Y, Sun, R, Zhou, Z.H. | | Deposit date: | 2017-09-25 | | Release date: | 2017-11-08 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure and mutagenesis reveal essential capsid protein interactions for KSHV replication.
Nature, 553, 2018
|
|
