8BCC
 
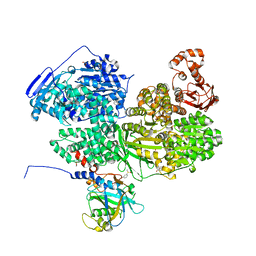 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 and compound 39 | | Descriptor: | 1,2-ETHANEDIOL, 3-oxidanylbenzenesulfonamide, Pre-mRNA-processing-splicing factor 8, ... | | Authors: | Vester, K, Loll, B, Wahl, M.C. | | Deposit date: | 2022-10-15 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Conformation-dependent ligand hot spots in the spliceosomal RNA helicase BRR2.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8BCB
 
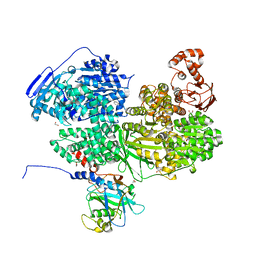 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 and compound 34 | | Descriptor: | 1,2-ETHANEDIOL, Pre-mRNA-processing-splicing factor 8, SULFANILAMIDE, ... | | Authors: | Vester, K, Loll, B, Wahl, M.C. | | Deposit date: | 2022-10-15 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Conformation-dependent ligand hot spots in the spliceosomal RNA helicase BRR2.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
6IH9
 
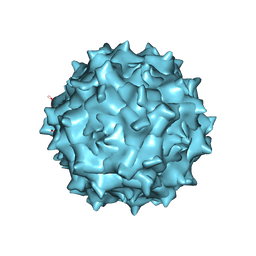 | | Adeno-Associated Virus 2 at 2.8 ang | | Descriptor: | Capsid protein VP1 | | Authors: | Lou, Z.Y, Zhang, R. | | Deposit date: | 2018-09-29 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Adeno-associated virus 2 bound to its cellular receptor AAVR.
Nat Microbiol, 4, 2019
|
|
6N51
 
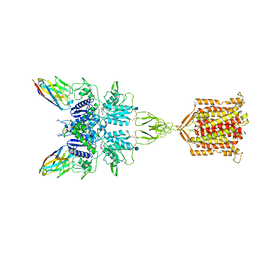 | | Metabotropic Glutamate Receptor 5 bound to L-quisqualate and Nb43 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 5, ... | | Authors: | Koehl, A, Hu, H, Feng, D, Sun, B, Weis, W.I, Skiniotis, G.S, Mathiesen, J.M, Kobilka, B.K. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into the activation of metabotropic glutamate receptors.
Nature, 566, 2019
|
|
8BCA
 
 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 and compound 26 | | Descriptor: | 1,2-ETHANEDIOL, 3-azanyl-~{N}-methyl-4-(methylamino)benzenesulfonamide, GLYCEROL, ... | | Authors: | Vester, K, Loll, B, Wahl, M.C. | | Deposit date: | 2022-10-15 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformation-dependent ligand hot spots in the spliceosomal RNA helicase BRR2.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7JP1
 
 | |
6T3I
 
 | | Solution structure of the HRP2 IBD | | Descriptor: | Hepatoma-derived growth factor-related protein 2 | | Authors: | Veverka, V. | | Deposit date: | 2019-10-11 | | Release date: | 2020-11-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Unlike Its Paralog LEDGF/p75, HRP-2 Is Dispensable for MLL-R Leukemogenesis but Important for Leukemic Cell Survival.
Cells, 10, 2021
|
|
8BCF
 
 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 and compound 78 | | Descriptor: | 1,2-ETHANEDIOL, Pre-mRNA-processing-splicing factor 8, U5 small nuclear ribonucleoprotein 200 kDa helicase, ... | | Authors: | Vester, K, Loll, B, Wahl, M.C. | | Deposit date: | 2022-10-15 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Conformation-dependent ligand hot spots in the spliceosomal RNA helicase BRR2.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8BCE
 
 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 and compound 76 | | Descriptor: | 1,2-ETHANEDIOL, N-methoxybenzenesulfonamide, Pre-mRNA-processing-splicing factor 8, ... | | Authors: | Vester, K, Loll, B, Wahl, M.C. | | Deposit date: | 2022-10-15 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conformation-dependent ligand hot spots in the spliceosomal RNA helicase BRR2.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7JPO
 
 | |
8BCG
 
 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 and compound 86 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-~{N}-methoxy-~{N}-methyl-benzenesulfonamide, Pre-mRNA-processing-splicing factor 8, ... | | Authors: | Vester, K, Loll, B, Wahl, M.C. | | Deposit date: | 2022-10-15 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Conformation-dependent ligand hot spots in the spliceosomal RNA helicase BRR2.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
6BX4
 
 | |
7JRO
 
 | | Plant Mitochondrial complex IV from Vigna radiata | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, CARDIOLIPIN, ... | | Authors: | Maldonado, M, Letts, J.A. | | Deposit date: | 2020-08-12 | | Release date: | 2021-01-20 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic structures of respiratory complex III 2 , complex IV, and supercomplex III 2 -IV from vascular plants.
Elife, 10, 2021
|
|
6N52
 
 | | Metabotropic Glutamate Receptor 5 Apo Form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 5 | | Authors: | Koehl, A, Hu, H, Feng, D, Sun, B, Weis, W.I, Skiniotis, G.S, Mathiesen, J.M, Kobilka, B.K. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into the activation of metabotropic glutamate receptors.
Nature, 566, 2019
|
|
8BC8
 
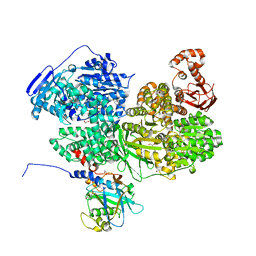 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 and compound 18 | | Descriptor: | 1,2-ETHANEDIOL, 3-azanyl-4-oxidanyl-benzenesulfonamide, Pre-mRNA-processing-splicing factor 8, ... | | Authors: | Vester, K, Loll, B, Wahl, M.C. | | Deposit date: | 2022-10-15 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Conformation-dependent ligand hot spots in the spliceosomal RNA helicase BRR2.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
6TCL
 
 | | Photosystem I tetramer | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Chen, M, Perez-Boerema, A, Li, S, Amunts, A. | | Deposit date: | 2019-11-06 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Distinct structural modulation of photosystem I and lipid environment stabilizes its tetrameric assembly.
Nat.Plants, 6, 2020
|
|
7JPQ
 
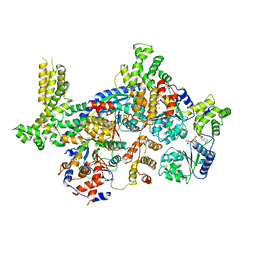 | | ORC-O2-5: Human Origin Recognition Complex (ORC) with subunits 2,3,4,5 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Origin recognition complex subunit 2, ... | | Authors: | Jaremko, M.J, Joshua-Tor, L. | | Deposit date: | 2020-08-09 | | Release date: | 2020-09-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The dynamic nature of the human Origin Recognition Complex revealed through five cryoEM structures.
Elife, 9, 2020
|
|
1A43
 
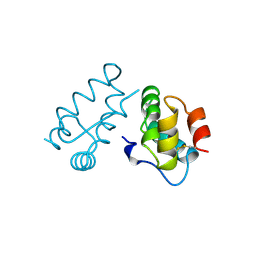 | | STRUCTURE OF THE HIV-1 CAPSID PROTEIN DIMERIZATION DOMAIN AT 2.6A RESOLUTION | | Descriptor: | HIV-1 CAPSID | | Authors: | Worthylake, D.K, Wang, H, Yoo, S, Sundquist, W.I, Hill, C.P. | | Deposit date: | 1998-02-10 | | Release date: | 1999-02-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the HIV-1 capsid protein dimerization domain at 2.6 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
8BCD
 
 | |
8FN9
 
 | |
6BMC
 
 | | The structure of a dimeric type II DAH7PS associated with pyocyanin biosynthesis in Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, COBALT (II) ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Sterritt, O.W, Jameson, G.B, Parker, E.J. | | Deposit date: | 2017-11-14 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional characterisation of the entry point to pyocyanin biosynthesis inPseudomonas aeruginosadefines a new 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase subclass.
Biosci. Rep., 38, 2018
|
|
7LMB
 
 | | Tetrahymena telomerase T5D5 structure at 3.8 Angstrom | | Descriptor: | Telomerase La-related protein p65, Telomerase RNA, Telomerase associated protein p50, ... | | Authors: | He, Y, Wang, Y, Liu, B, Helmling, C, Susac, L, Cheng, R, Zhou, Z.H, Feigon, J. | | Deposit date: | 2021-02-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of telomerase at several steps of telomere repeat synthesis.
Nature, 593, 2021
|
|
7LMO
 
 | | Structure of full-length human lambda-6A light chain JTO in complex with stabilizer 34 [3-(2-(7-(diethylamino)-4-methyl-2-oxo-2H-chromen-3-yl)ethyl)-7-(1H-imidazole-5-carbonyl)-1,3,7-triazaspiro[4.4]nonane-2,4-dione] | | Descriptor: | (5~{R})-3-[2-[7-(diethylamino)-4-methyl-2-oxidanylidene-chromen-3-yl]ethyl]-7-(1~{H}-imidazol-4-ylcarbonyl)-1,3,7-triazaspiro[4.4]nonane-2,4-dione, JTO light chain, PHOSPHATE ION | | Authors: | Yan, N.L, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2021-02-05 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of Potent Coumarin-Based Kinetic Stabilizers of Amyloidogenic Immunoglobulin Light Chains Using Structure-Based Design.
J.Med.Chem., 64, 2021
|
|
7AZ5
 
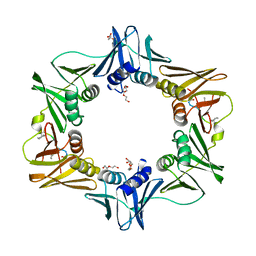 | | DNA polymerase sliding clamp from Escherichia coli with peptide 47 bound | | Descriptor: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, Peptide 47, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7LMN
 
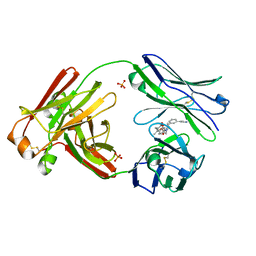 | |
