8VRG
 
 | | E. coli peptidyl-prolyl cis-trans isomerase containing delta1-monofluoro-leucines | | Descriptor: | Peptidyl-prolyl cis-trans isomerase B | | Authors: | Frkic, R.L, Jackson, C.J. | | Deposit date: | 2024-01-22 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Conformational Preferences of the Non-Canonical Amino Acids (2 S ,4 S )-5-Fluoroleucine, (2 S ,4 R )-5-Fluoroleucine, and 5,5'-Difluoroleucine in a Protein.
Biochemistry, 63, 2024
|
|
8VRE
 
 | | Structure of PYCR1 complexed with NADH and N-formyl-L-proline | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1-formyl-L-proline, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2024-01-21 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1)
Protein Sci., 33, 2024
|
|
8VRD
 
 | |
8VR7
 
 | | crystal structure of the Pcryo_0619 N-acetyltransferase from Psychrobacter cryohalolentis K5 int he presence of acetyl coenzyme A | | Descriptor: | 1,2-ETHANEDIOL, 3'-PHOSPHATE-ADENOSINE-5'-DIPHOSPHATE, 3-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]PROPANE-1-SULFONIC ACID, ... | | Authors: | Dunsirn, M.M, Bockhaus, N.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2024-01-20 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical Investigation of the Enzymes Required for the Production of 2,3,4-triacetoamido-2,3,4-trideoxy-l-arabinose in Psychrobacter cryohalolentis K5
To Be Published
|
|
8VR6
 
 | | crystal structure of the Pcryo_0619 N-acetryltransferase from Psychrobacter cryohalolentis K5 in the presence of CoA-disulfide | | Descriptor: | Acetyltransferase, putative, [[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3~{R})-4-[[3-[2-[2-[3-[[(2~{R})-4-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyldisulfanyl]ethylamino]-3-oxidanylidene-propyl]amino]-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-butyl] hydrogen phosphate | | Authors: | Dunsirn, M.M, Bockhaus, N.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2024-01-20 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical Investigation of the Enzymes Required for the Production of 2,3,4-triacetoamido-2,3,4-trideoxy-l-arabinose in Psychrobacter cryohalolentis K5
To Be Published
|
|
8VR5
 
 | | crystal structure of the Pcryo_0618 aminotransferase from Psychrobacter cryohalolentis K5 in the presence of PMP and glutamate | | Descriptor: | 1,2-ETHANEDIOL, 2,2'-(1,4-diazepane-1,4-diyl)di(ethane-1-sulfonic acid), 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Bockhaus, N.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2024-01-20 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical Investigation of the Enzymes Required for the Production of 2,3,4-triacetoamido-2,3,4-trideoxy-l-arabinose in Psychrobacter cryohalolentis K5
To Be Published
|
|
8VR3
 
 | | crystal structure of the Pcryo_0618 aminotransferase from Psychrobacter cryohalolentis K5 in the presence of its internal aldimine | | Descriptor: | 1,2-ETHANEDIOL, 2,2'-(1,4-diazepane-1,4-diyl)di(ethane-1-sulfonic acid), CHLORIDE ION, ... | | Authors: | Bockhaus, N.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2024-01-20 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical Investigation of the Enzymes Required for the Production of 2,3,4-triacetoamido-2,3,4-trideoxy-l-arabinose in Psychrobacter cryohalolentis K5
To Be Published
|
|
8VR2
 
 | | Crystal structure of the Pcryo_0617 oxidoreductase/decarboxylase from Psychrobacter cryohalolentis K5 in the presence of NAD and UDP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NAD-dependent epimerase/dehydratase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Bockhaus, N.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2024-01-20 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical Investigation of the Enzymes Required for the Production of 2,3,4-triacetoamido-2,3,4-trideoxy-l-arabinose in Psychrobacter cryohalolentis K5
To Be Published
|
|
8VR1
 
 | |
8VR0
 
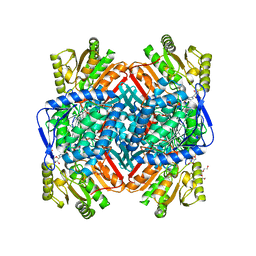 | |
8VQZ
 
 | |
8VQY
 
 | | Human GABAA receptor alpha1-beta2-gamma2 subtype in complex with GABA plus methaqualone | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-methyl-3-(2-methylphenyl)quinazolin-4(3H)-one, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Chojnacka, W, Teng, J, Kim, J.J, Jensen, A.A, Hibbs, R.E. | | Deposit date: | 2024-01-19 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Structural insights into GABA A receptor potentiation by Quaalude.
Nat Commun, 15, 2024
|
|
8VQW
 
 | |
8VQR
 
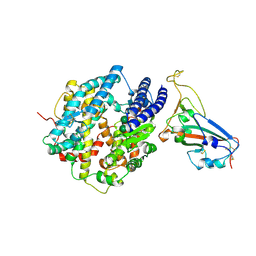 | | Crystal structure of chimeric SARS-CoV-2 RBD complexed with chimeric raccoon dog ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsueh, F.-C, Shi, K, Aihara, H, Li, F. | | Deposit date: | 2024-01-19 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.565 Å) | | Cite: | Structural basis for raccoon dog receptor recognition by SARS-CoV-2
To Be Published
|
|
8VQQ
 
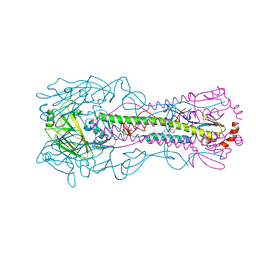 | | Crystal structure of the A/Puerto Rico/8/1934 (H1N1) influenza virus hemagglutinin in complex with small molecule 6S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain, ... | | Authors: | Lin, T.H, Zhu, Y, Wilson, I.A. | | Deposit date: | 2024-01-19 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Ultrapotent influenza hemagglutinin fusion inhibitors developed through SuFEx-enabled high-throughput medicinal chemistry.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VQN
 
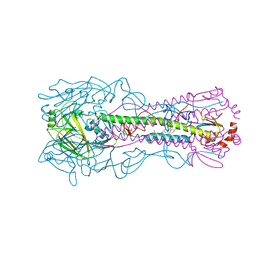 | | Crystal structure of the A/Puerto Rico/8/1934 (H1N1) influenza virus hemagglutinin in complex with small molecule 6R | | Descriptor: | (S~1~S,3R)-N-{3-chloro-4-[(2S)-2-phenylmorpholine-4-carbonyl]phenyl}-3-(dimethylamino)pyrrolidine-1-sulfonimidoyl fluoride, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Lin, T.H, Zhu, Y, Wilson, I.A. | | Deposit date: | 2024-01-18 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Ultrapotent influenza hemagglutinin fusion inhibitors developed through SuFEx-enabled high-throughput medicinal chemistry.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VQM
 
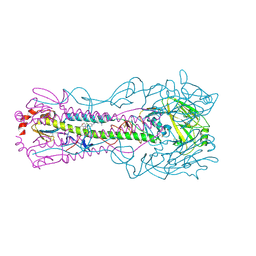 | | Crystal structure of the A/Puerto Rico/8/1934 (H1N1) influenza virus hemagglutinin in complex with small molecule 6R prime | | Descriptor: | (S~1~S,3R)-N-{3,5-dichloro-4-[(2S)-2-phenylmorpholine-4-carbonyl]phenyl}-3-(dimethylamino)pyrrolidine-1-sulfonimidoyl fluoride, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lin, T.H, Zhu, Y, Wilson, I.A. | | Deposit date: | 2024-01-18 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Ultrapotent influenza hemagglutinin fusion inhibitors developed through SuFEx-enabled high-throughput medicinal chemistry.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VQL
 
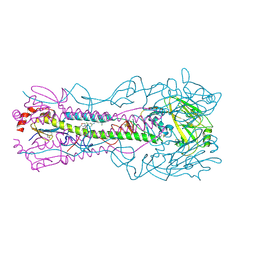 | | Crystal structure of the A/Puerto Rico/8/1934 (H1N1) influenza virus hemagglutinin in complex with small molecule 6S prime | | Descriptor: | (S~1~S,3S)-N-{3,5-dichloro-4-[(2S)-2-phenylmorpholine-4-carbonyl]phenyl}-3-(dimethylamino)pyrrolidine-1-sulfonimidoyl fluoride, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lin, T.H, Zhu, Y, Wilson, I.A. | | Deposit date: | 2024-01-18 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Ultrapotent influenza hemagglutinin fusion inhibitors developed through SuFEx-enabled high-throughput medicinal chemistry.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VQ4
 
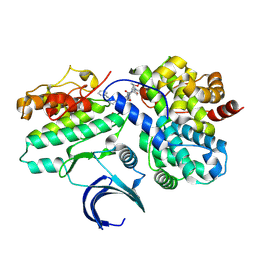 | | CDK2-CyclinE1 in complex with allosteric inhibitor I-125A. | | Descriptor: | (8R)-6-(1-benzyl-1H-pyrazole-4-carbonyl)-N-[(2S,3R)-3-(2-cyclohexylethoxy)-1-(methylamino)-1-oxobutan-2-yl]-2-[(1S)-2,2-dimethylcyclopropane-1-carbonyl]-2,6-diazaspiro[3.4]octane-8-carboxamide, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | Hirschi, M, Johnson, E, Zhang, Y, Liu, Z, Brodsky, O, Won, S.J, Nagata, A, Petroski, M.D, Majmudar, J.D, Niessen, S, VanArsdale, T, Gilbert, A.M, Hayward, M.M, Stewart, A.E, Nager, A.R, Melillo, B, Cravatt, B. | | Deposit date: | 2024-01-17 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Expanding the ligandable proteome by paralog hopping with covalent probes.
Biorxiv, 2024
|
|
8VQ3
 
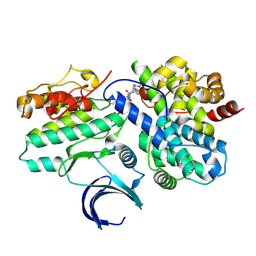 | | CDK2-CyclinE1 in complex with allosteric inhibitor I-198. | | Descriptor: | (8R)-N-[(2S,3R)-3-(cyclohexylmethoxy)-1-(morpholin-4-yl)-1-oxobutan-2-yl]-2-[(1S)-2,2-dimethylcyclopropane-1-carbonyl]-6-(1,3-thiazole-5-carbonyl)-2,6-diazaspiro[3.4]octane-8-carboxamide, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | Hirschi, M, Johnson, E, Zhang, Y, Liu, Z, Brodsky, O, Won, S.J, Nagata, A, Petroski, M.D, Majmudar, J.D, Niessen, S, VanArsdale, T, Gilbert, A.M, Hayward, M.M, Stewart, A.E, Nager, A.R, Melillo, B, Cravatt, B. | | Deposit date: | 2024-01-17 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Expanding the ligandable proteome by paralog hopping with covalent probes.
Biorxiv, 2024
|
|
8VQ1
 
 | | Pseudomonas fluorescens G150T isocyanide hydratase at 298 K XFEL data, thioimidate intermediate | | Descriptor: | CHLORIDE ION, Isonitrile hydratase InhA, N-(4-nitrophenyl)methanimine | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2024-01-17 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Changes in an enzyme ensemble during catalysis observed by high-resolution XFEL crystallography.
Sci Adv, 10, 2024
|
|
8VPW
 
 | | Pseudomonas fluorescens G150T isocyanide hydratase at 298 K XFEL data, free enzyme | | Descriptor: | CHLORIDE ION, Isonitrile hydratase InhA | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2024-01-17 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Changes in an enzyme ensemble during catalysis observed by high-resolution XFEL crystallography.
Sci Adv, 10, 2024
|
|
8VPO
 
 | | X-Ray Crystal Structure of TigE from Paramaledivibacter caminithermalis | | Descriptor: | GLYCEROL, IRON/SULFUR CLUSTER, Radical SAM core domain-containing protein | | Authors: | Grove, T.L, Lachowicz, J.C, Zizola, C. | | Deposit date: | 2024-01-16 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural, Biochemical, and Bioinformatic Basis for Identifying Radical SAM Cyclopropyl Synthases.
Acs Chem.Biol., 19, 2024
|
|
8VOV
 
 | | Structure of VCP in complex with an ATPase activator and ADP (D2 domains only, hexameric form) | | Descriptor: | (3R)-N-[2-(ethylsulfanyl)phenyl]-3-(1-oxo-1,3-dihydro-2H-isoindol-2-yl)butanamide, ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Jones, N.H, Urnivicius, L, Kapoor, T.M. | | Deposit date: | 2024-01-16 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Allosteric activation of VCP, an AAA unfoldase, by small molecule mimicry.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VOI
 
 | |
