6U0P
 
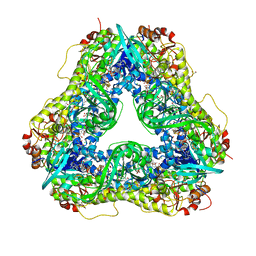 | | Crystal structure of PieE, the flavin-dependent monooxygenase involved in the biosynthesis of piericidin A1 | | Descriptor: | 2,4-dichlorophenol 6-monooxygenase, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Shi, R, Manenda, M, Picard, M.-E. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural analyses of the Group A flavin-dependent monooxygenase PieE reveal a sliding FAD cofactor conformation bridging OUT and IN conformations.
J.Biol.Chem., 295, 2020
|
|
6UI5
 
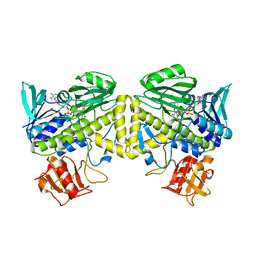 | |
2DKI
 
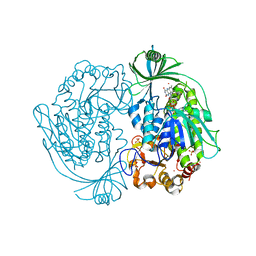 | | Crystal structure of 3-hydroxybenzoate hydroxylase from Comamonas testosteroni, under pressure of xenon gas (12 atm) | | Descriptor: | 3-HYDROXYBENZOATE HYDROXYLASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Hiromoto, T, Fujiwara, S, Hosokawa, K, Yamaguchi, H. | | Deposit date: | 2006-04-11 | | Release date: | 2006-10-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of 3-hydroxybenzoate hydroxylase from Comamonas testosteroni has a large tunnel for substrate and oxygen access to the active site
J.Mol.Biol., 364, 2006
|
|
4EIP
 
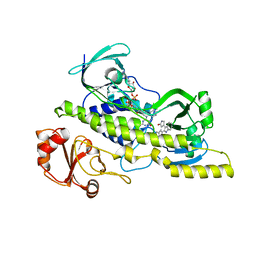 | | Native and K252c bound RebC-10x | | Descriptor: | 6,7,12,13-tetrahydro-5H-indolo[2,3-a]pyrrolo[3,4-c]carbazol-5-one, FLAVIN-ADENINE DINUCLEOTIDE, Putative FAD-monooxygenase | | Authors: | Goldman, P.J, Ryan, K.S, Howard-Jones, A.R, Hamill, M.J, Elliott, S.J, Walsh, C.T, Drennan, C.L. | | Deposit date: | 2012-04-05 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.332 Å) | | Cite: | An Unusual Role for a Mobile Flavin in StaC-like Indolocarbazole Biosynthetic Enzymes.
Chem.Biol., 19, 2012
|
|
5XGV
 
 | |
4EIQ
 
 | | Chromopyrrolic acid-soaked RebC-10x with bound 7-carboxy-K252c | | Descriptor: | (5S)-7-oxo-6,7,12,13-tetrahydro-5H-indolo[2,3-a]pyrrolo[3,4-c]carbazole-5-carboxylic acid, Putative FAD-monooxygenase | | Authors: | Goldman, P.J, Ryan, K.S, Howard-Jones, A.R, Hamill, M.J, Elliott, S.J, Walsh, C.T, Drennan, C.L. | | Deposit date: | 2012-04-05 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | An Unusual Role for a Mobile Flavin in StaC-like Indolocarbazole Biosynthetic Enzymes.
Chem.Biol., 19, 2012
|
|
2QA1
 
 | | Crystal structure of PgaE, an aromatic hydroxylase involved in angucycline biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Koskiniemi, H, Dobritzsch, D, Metsa-Ketela, M, Kallio, P, Niemi, J, Schneider, G. | | Deposit date: | 2007-06-14 | | Release date: | 2007-08-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of two aromatic hydroxylases involved in the early tailoring steps of angucycline biosynthesis
J.Mol.Biol., 372, 2007
|
|
2QA2
 
 | | Crystal structure of CabE, an aromatic hydroxylase from angucycline biosynthesis, determined to 2.7 A resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Polyketide oxygenase CabE | | Authors: | Koskiniemi, H, Dobritzsch, D, Metsa-Ketela, M, Kallio, P, Niemi, J, Schneider, G. | | Deposit date: | 2007-06-14 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of two aromatic hydroxylases involved in the early tailoring steps of angucycline biosynthesis
J.Mol.Biol., 372, 2007
|
|
2R0C
 
 | |
4Z2U
 
 | |
5VQB
 
 | | Crystal structure of rifampin monooxygenase from Streptomyces venezuelae, complex with FAD | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Cox, G, Kelso, J, Stogios, P.J, Savchenko, A, Anderson, W.F, Wright, G.D, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-05-08 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.391 Å) | | Cite: | Rox, a Rifamycin Resistance Enzyme with an Unprecedented Mechanism of Action.
Cell Chem Biol, 25, 2018
|
|
4Z2R
 
 | |
4Z2T
 
 | |
2VOU
 
 | | Structure of 2,6-dihydroxypyridine-3-hydroxylase from Arthrobacter nicotinovorans | | Descriptor: | 2,6-DIHYDROXYPYRIDINE HYDROXYLASE, ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Treiber, N, Schulz, G.E. | | Deposit date: | 2008-02-21 | | Release date: | 2008-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of 2,6-Dihydroxypyridine 3-Hydroxylase from a Nicotine-Degrading Pathway.
J.Mol.Biol., 379, 2008
|
|
5BRT
 
 | | Crystal Structure of 2-hydroxybiphenyl 3-monooxygenase from Pseudomonas azelaica with 2-hydroxybiphenyl in the active site | | Descriptor: | 2-HYDROXYBIPHENYL, 2-hydroxybiphenyl-3-monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kanteev, M, Bregman-Cohen, A, Deri, B, Adir, N, Fishman, A. | | Deposit date: | 2015-06-01 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A crystal structure of 2-hydroxybiphenyl 3-monooxygenase with bound substrate provides insights into the enzymatic mechanism.
Biochim.Biophys.Acta, 1854, 2015
|
|
2R0G
 
 | | Chromopyrrolic acid-soaked RebC with bound 7-carboxy-K252c | | Descriptor: | 7-carboxy-5-hydroxy-12,13-dihydro-6H-indolo[2,3-a]pyrrolo[3,4-c]carbazole, FLAVIN-ADENINE DINUCLEOTIDE, RebC | | Authors: | Ryan, K.S, Drennan, C.L. | | Deposit date: | 2007-08-19 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystallographic trapping in the rebeccamycin biosynthetic enzyme RebC
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2R0P
 
 | | K252c-soaked RebC | | Descriptor: | 6,7,12,13-tetrahydro-5H-indolo[2,3-a]pyrrolo[3,4-c]carbazol-5-one, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Ryan, K.S, Drennan, C.L. | | Deposit date: | 2007-08-20 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic trapping in the rebeccamycin biosynthetic enzyme RebC
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1FOH
 
 | | PHENOL HYDROXYLASE FROM TRICHOSPORON CUTANEUM | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHENOL, PHENOL HYDROXYLASE | | Authors: | Enroth, C, Neujahr, H, Schneider, G, Lindqvist, Y. | | Deposit date: | 1998-03-26 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of phenol hydroxylase in complex with FAD and phenol provides evidence for a concerted conformational change in the enzyme and its cofactor during catalysis.
Structure, 6, 1998
|
|
1PN0
 
 | | Phenol hydroxylase from Trichosporon cutaneum | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, PHENOL, ... | | Authors: | Enroth, C. | | Deposit date: | 2003-06-12 | | Release date: | 2003-09-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution structure of phenol hydroxylase and correction of sequence errors.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
4HB9
 
 | |
4K2X
 
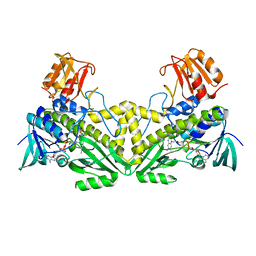 | |
4K5R
 
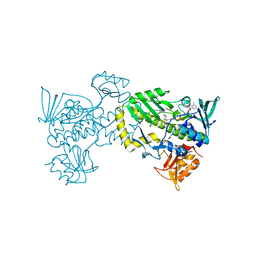 | | The 2.0 angstrom crystal structure of MTMOIV, a baeyer-villiger monooxygenase from the mithramycin biosynthetic pathway in streptomyces argillaceus. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Oxygenase | | Authors: | Noinaj, N, Bosserman, M.A, Rohr, J, Buchanan, S.K. | | Deposit date: | 2013-04-15 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Insight into Substrate Recognition and Catalysis of Baeyer-Villiger Monooxygenase MtmOIV, the Key Frame-Modifying Enzyme in the Biosynthesis of Anticancer Agent Mithramycin.
Acs Chem.Biol., 8, 2013
|
|
4K5S
 
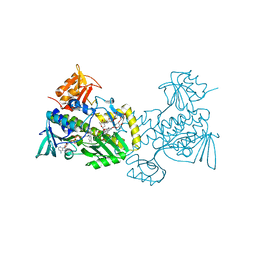 | | The crystal structure of premithramycin B in complex with MTMOIV, a baeyer-villiger monooxygenase from the mithramycin biosynthetic pathway in streptomyces argillaceus. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Oxygenase, premithramycin B | | Authors: | Noinaj, N, Bosserman, M.A, Rohr, J, Buchanan, S.K. | | Deposit date: | 2013-04-15 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Insight into Substrate Recognition and Catalysis of Baeyer-Villiger Monooxygenase MtmOIV, the Key Frame-Modifying Enzyme in the Biosynthesis of Anticancer Agent Mithramycin.
Acs Chem.Biol., 8, 2013
|
|
4ICY
 
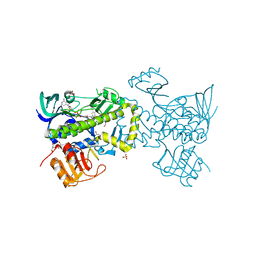 | | Tracing the Evolution of Angucyclinone Monooxygenases: Structural Determinants for C-12b Hydroxylation and Substrate Inhibition in PgaE | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kallio, P, Patrikainen, P, Belogurov, G, Mantsala, P, Yang, K, Niemi, J, Metsa-Ketela, M. | | Deposit date: | 2012-12-11 | | Release date: | 2013-06-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Tracing the Evolution of Angucyclinone Monooxygenases: Structural Determinants for C-12b Hydroxylation and Substrate Inhibition in PgaE.
Biochemistry, 52, 2013
|
|
6ICI
 
 | | Crystal structure of human MICAL3 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, [F-actin]-monooxygenase MICAL3 | | Authors: | Hwang, K.Y, Kim, J.S. | | Deposit date: | 2018-09-06 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic insights into flavin-containing monooxygenase and calponin-homology domains in human MICAL3.
Iucrj, 7, 2020
|
|
