2EIJ
 
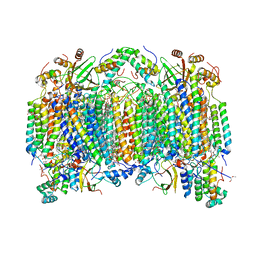 | | Bovine heart cytochrome C oxidase in the fully reduced state | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Muramoto, K, Hirata, K, Shinzawa-Itoh, K, Yoko-o, S, Yamashita, E, Aoyama, H, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2007-03-13 | | Release date: | 2007-05-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A histidine residue acting as a controlling site for dioxygen reduction and proton pumping by cytochrome c oxidase
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
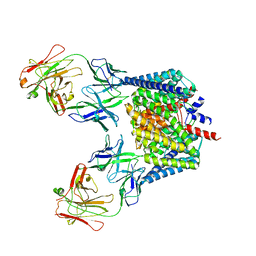
2H2S
 
 | |
7VSO
 
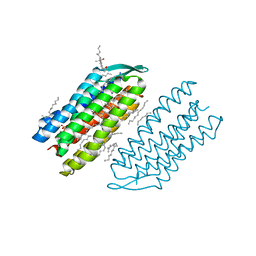 | | Serial Femtosecond Crystallography (SFX) of Ground State Bacteriorhodopsin Crystallized from Bicelles in Complex with HAD16 Determined Using 7-keV X-ray Free Electron Laser (XFEL) at SACLA | | Descriptor: | 2-[[(2R)-2-(3-bromanyl-5-iodanyl-phenyl)carbonyloxy-3-tetradecanoyloxy-propoxy]-oxidanyl-phosphoryl]oxyethyl-trimethyl-azanium, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Bacteriorhodopsin, ... | | Authors: | Mizohata, E, Nakane, T, Hanashima, S. | | Deposit date: | 2021-10-27 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Heavy Atom Detergent/Lipid Combined X-ray Crystallography for Elucidating the Structure-Function Relationships of Membrane Proteins.
Membranes (Basel), 11, 2021
|
|
2E6M
 
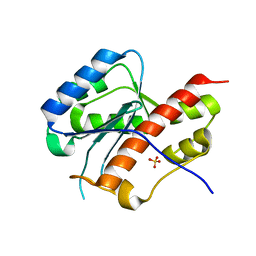 | | structure of mouse werner exonuclease domain | | Descriptor: | SULFATE ION, Werner syndrome ATP-dependent helicase homolog | | Authors: | Cho, Y, Choi, J.M. | | Deposit date: | 2006-12-27 | | Release date: | 2007-01-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | probing the roles of active site residues in 3'-5' exonuclease of werner syndrome protein
TO BE PUBLISHED
|
|
7V33
 
 | | Active state complex I from rotenone-NADH dataset | | Descriptor: | (2R,6aS,12aS)-8,9-dimethoxy-2-(prop-1-en-2-yl)-1,2,12,12a-tetrahydrofuro[2',3':7,8][1]benzopyrano[2,3-c][1]benzopyran-6(6aH)-one, (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-10 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7V2D
 
 | | Deactive state complex I from Q10 dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-08 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7VEK
 
 | | Crystal structure of Phytolacca americana UGT3 with capsaicin and UDP-2fluoroglucose | | Descriptor: | (6E)-N-(4-hydroxy-3-methoxybenzyl)-8-methylnon-6-enamide, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, Glycosyltransferase, ... | | Authors: | Maharjan, R, Fukuda, Y, Nakayama, T, Nakayama, T, Hamada, H, Ozaki, S, Inoue, T. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for substrate recognition in the Phytolacca americana glycosyltransferase PaGT3.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7V2R
 
 | | Active state complex I from Q1-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-09 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
2H8P
 
 | | Structure of a K channel with an amide to ester substitution in the selectivity filter | | Descriptor: | (2S)-2-(BUTYRYLOXY)-3-HYDROXYPROPYL NONANOATE, FAB heavy chain, FAB light chain, ... | | Authors: | Valiyaveetil, F.I, MacKinnon, R, Muir, T.W. | | Deposit date: | 2006-06-07 | | Release date: | 2006-09-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and Functional Consequences of an Amide-to-Ester Substitution in the Selectivity Filter of a Potassium Channel.
J.Am.Chem.Soc., 128, 2006
|
|
7V2K
 
 | | Deactive state complex I from DQ-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-09 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7VEL
 
 | | Crystal structure of Phytolacca americana UGT3 with UDP-2fluoroglucose | | Descriptor: | 1,2-ETHANEDIOL, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, Glycosyltransferase, ... | | Authors: | Maharjan, R, Fukuda, Y, Nakayama, T, Nakayama, T, Hamada, H, Ozaki, S, Inoue, T. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for substrate recognition in the Phytolacca americana glycosyltransferase PaGT3.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7V2E
 
 | | Active state complex I from Q10-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-08 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7VEJ
 
 | | Crystal structure of Phytolacca americana UGT3 with kaempferol and UDP-2fluoroglucose | | Descriptor: | 1,2-ETHANEDIOL, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 3,5,7-TRIHYDROXY-2-(4-HYDROXYPHENYL)-4H-CHROMEN-4-ONE, ... | | Authors: | Maharjan, R, Fukuda, Y, Nakayama, T, Nakayama, T, Hamada, H, Ozaki, S, Inoue, T. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for substrate recognition in the Phytolacca americana glycosyltransferase PaGT3.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7V2F
 
 | | Deactive state complex I from Q10-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-09 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7V2H
 
 | | Active state complex I from DQ-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-09 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
2GLX
 
 | | Crystal Structure Analysis of bacterial 1,5-AF Reductase | | Descriptor: | 1,5-anhydro-D-fructose reductase, ACETATE ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Dambe, T.R, Scheidig, A.J. | | Deposit date: | 2006-04-05 | | Release date: | 2006-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of NADP(H)-Dependent 1,5-Anhydro-d-fructose Reductase from Sinorhizobium morelense at 2.2 A Resolution: Construction of a NADH-Accepting Mutant and Its Application in Rare Sugar Synthesis
Biochemistry, 45, 2006
|
|
2ERJ
 
 | | Crystal structure of the heterotrimeric interleukin-2 receptor in complex with interleukin-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Debler, E.W, Stauber, D.J, Wilson, I.A. | | Deposit date: | 2005-10-25 | | Release date: | 2006-02-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the IL-2 signaling complex: Paradigm for a heterotrimeric cytokine receptor.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HBJ
 
 | | Structure of the yeast nuclear exosome component, Rrp6p, reveals an interplay between the active site and the HRDC domain | | Descriptor: | Exosome complex exonuclease RRP6 | | Authors: | Midtgaard, S.F, Assenholt, J, Jonstrup, A.T, Van, L.B, Jensen, T.H, Brodersen, D.E. | | Deposit date: | 2006-06-14 | | Release date: | 2006-07-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the nuclear exosome component Rrp6p reveals an interplay between the active site and the HRDC domain.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
7V30
 
 | | Deactive state complex I from Q1-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-10 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TOF
 
 | | Structure of G6PD-WT dimer with no symmetry applied | | Descriptor: | Glucose-6-phosphate 1-dehydrogenase | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2022-01-24 | | Release date: | 2022-09-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Allosteric role of a structural NADP + molecule in glucose-6-phosphate dehydrogenase activity.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2HE7
 
 | | FERM domain of EPB41L3 (DAL-1) | | Descriptor: | Band 4.1-like protein 3 | | Authors: | Hallberg, B.M, Busam, R.D, Arrowsmith, C, Berglund, H, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Schiavone, L.H, Johansson, I, Hogbom, M, Karlberg, T, Kotenyova, T, Nilvebrandt, J, Norberg, P, Stenmark, P, Nordlund, P, Nilsson-ehle, P, Nyman, T, Ogg, D, Sagemark, J, Sundstrom, M, Uppenberg, J, Van den berg, S, Weigelt, J, Persson, C, Thorsell, A.G, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-21 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of tumor suppressor in lung cancer 1 (TSLC1) binding to differentially expressed in adenocarcinoma of the lung (DAL-1/4.1B).
J.Biol.Chem., 286, 2011
|
|
2HF4
 
 | | Crystal structure of Monomeric Actin in its ATP-bound state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-5C, CALCIUM ION | | Authors: | Rould, M.A, Wan, Q, Joel, P.B, Lowey, S, Trybus, K.M. | | Deposit date: | 2006-06-22 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Expressed Non-polymerizable Monomeric Actin in the ADP and ATP States.
J.Biol.Chem., 281, 2006
|
|
2GST
 
 | | STRUCTURE OF THE XENOBIOTIC SUBSTRATE BINDING SITE OF A GLUTATHIONE S-TRANSFERASE AS REVEALED BY X-RAY CRYSTALLOGRAPHIC ANALYSIS OF PRODUCT COMPLEXES WITH THE DIASTEREOMERS OF 9-(S-GLUTATHIONYL)-10-HYDROXY-9, 10-DIHYDROPHENANTHRENE | | Descriptor: | GLUTATHIONE S-TRANSFERASE, L-gamma-glutamyl-S-[(9S,10S)-10-hydroxy-9,10-dihydrophenanthren-9-yl]-L-cysteinylglycine, SULFATE ION | | Authors: | Ji, X, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1993-06-07 | | Release date: | 1993-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and function of the xenobiotic substrate binding site of a glutathione S-transferase as revealed by X-ray crystallographic analysis of product complexes with the diastereomers of 9-(S-glutathionyl)-10-hydroxy-9,10-dihydrophenanthrene.
Biochemistry, 33, 1994
|
|
2GIV
 
 | | Human MYST histone acetyltransferase 1 | | Descriptor: | ACETYL COENZYME *A, CHLORIDE ION, Probable histone acetyltransferase MYST1, ... | | Authors: | Min, J, Wu, H, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-03-29 | | Release date: | 2006-04-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The Crystal Structure of Human MYST histone acetyltransferase 1 in complex with acetylcoenzyme A
To be Published
|
|
7TOE
 
 | |
