8CYG
 
 | |
8RU3
 
 | | Crystal structure of beta-catenin in complex with alpha-helical peptide inhibitor | | Descriptor: | Axin-1, CHLORIDE ION, Catenin beta-1 | | Authors: | Yeste Vazquez, A, Klintrot, C.I.R, Grossmann, T.N, Hennig, S. | | Deposit date: | 2024-01-30 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structure-Based Design of Bicyclic Helical Peptides That Target the Oncogene beta-Catenin.
Angew.Chem.Int.Ed.Engl., 2024
|
|
6V0L
 
 | | PDGFR-b Promoter Forms a G-Vacancy Quadruplex that Can be Complemented by dGMP: Molecular Structure and Recognition of Guanine Derivatives and Metabolites | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, DNA (5'-D(*(3D1)P*AP*GP*GP*GP*AP*GP*GP*GP*CP*GP*GP*CP*GP*GP*GP*AP*CP*A)-3') | | Authors: | Wang, K.B, Dickerhoff, J, Wu, G, Yang, D. | | Deposit date: | 2019-11-18 | | Release date: | 2020-03-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | PDGFR-beta Promoter Forms a Vacancy G-Quadruplex that Can Be Filled in by dGMP: Solution Structure and Molecular Recognition of Guanine Metabolites and Drugs.
J.Am.Chem.Soc., 142, 2020
|
|
1C0O
 
 | |
6UZ0
 
 | | Cardiac sodium channel with flecainide | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jiang, D, Shi, H, Tonggu, L, Lenaeus, M.J, Zheng, N, Catterall, W.A. | | Deposit date: | 2019-11-14 | | Release date: | 2020-01-01 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structure of the Cardiac Sodium Channel.
Cell, 180, 2020
|
|
6V1I
 
 | | Cryo-EM reconstruction of the thermophilic bacteriophage P74-26 small terminase- symmetric | | Descriptor: | Small terminase protein | | Authors: | Hayes, J.A, Hilbert, B.J, Gaubitz, C, Stone, N.P, Kelch, B.A. | | Deposit date: | 2019-11-20 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A thermophilic phage uses a small terminase protein with a fixed helix-turn-helix geometry.
J.Biol.Chem., 295, 2020
|
|
5IRT
 
 | |
2BY4
 
 | | SR Ca(2+)-ATPase in the HnE2 state complexed with the thapsigargin derivative Boc-12ADT. | | Descriptor: | (3S,3AR,4S,6S,6AR,7S,8S,9BS)-6-(ACETYLOXY)-3,3A-DIHYDROXY-3,6,9-TRIMETHYL-8-{[(2Z)-2-METHYLBUT-2-ENOYL]OXY}-7-(OCTANOYLOXY)-2-OXO-2,3,3A,4,5,6,6A,7,8,9B-DECAHYDROAZULENO[4,5-B]FURAN-4-YL 12-[(TERT-BUTOXYCARBONYL)AMINO]DODECANOATE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Jensen, A.L, Moller, J.V, Soehoel, H, Denmeade, S.R, Isaacs, J.T, Olsen, C.E, Christensen, S.B, Nissen, P. | | Deposit date: | 2005-07-28 | | Release date: | 2006-04-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Natural Products as Starting Materials for Development of Second-Generation Serca Inhibitors Targeted Towards Prostate Cancer Cells
Bioorg.Med.Chem., 14, 2006
|
|
6PPT
 
 | |
6VCC
 
 | | Cryo-EM structure of the Dvl2 DIX filament | | Descriptor: | Segment polarity protein dishevelled homolog DVL-2 | | Authors: | Enos, M, Kan, W, Muennich, S, Chen, D.H, Skiniotis, G, Weis, W.I. | | Deposit date: | 2019-12-20 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Limited Dishevelled/Axin oligomerization determines efficiency of Wnt/ beta-catenin signal transduction.
Elife, 9, 2020
|
|
1BTO
 
 | |
5J7J
 
 | |
1C0G
 
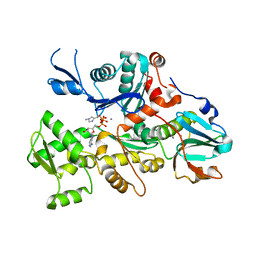 | | CRYSTAL STRUCTURE OF 1:1 COMPLEX BETWEEN GELSOLIN SEGMENT 1 AND A DICTYOSTELIUM/TETRAHYMENA CHIMERA ACTIN (MUTANT 228: Q228K/T229A/A230Y/E360H) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, PROTEIN (CHIMERIC ACTIN), ... | | Authors: | Matsuura, Y, Stewart, M, Kawamoto, M, Kamiya, N, Saeki, K, Yasunaga, T, Wakabayashi, T. | | Deposit date: | 1999-07-16 | | Release date: | 2000-03-01 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the higher Ca(2+)-activation of the regulated actin-activated myosin ATPase observed with Dictyostelium/Tetrahymena actin chimeras.
J.Mol.Biol., 296, 2000
|
|
1BTL
 
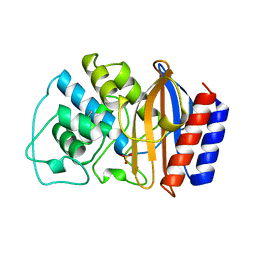 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI TEM1 BETA-LACTAMASE AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | BETA-LACTAMASE TEM1, SULFATE ION | | Authors: | Jelsch, C, Mourey, L, Masson, J.M, Samama, J.P. | | Deposit date: | 1993-11-01 | | Release date: | 1995-01-26 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Escherichia coli TEM1 beta-lactamase at 1.8 A resolution.
Proteins, 16, 1993
|
|
6VAO
 
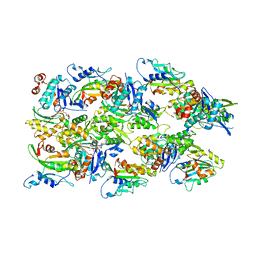 | | Human cofilin-1 decorated actin filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Huehn, A.R, Bibeau, J.P, Schramm, A.C, Cao, W, De La Cruz, E.M, Sindelar, C.V. | | Deposit date: | 2019-12-17 | | Release date: | 2020-01-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of cofilin-induced structural changes reveal local and asymmetric perturbations of actin filaments.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8WKQ
 
 | | Cryo-EM structure of the MS ring (C1) with export apparatus and proximal rod within the flagellar motor-hook complex in the CW state. | | Descriptor: | Flagellar M-ring protein, Flagellar basal body rod protein FlgB, Flagellar basal-body rod protein FlgC, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-09-28 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the MS ring (C1) with export apparatus and proximal rod within the flagellar motor-hook complex in the CW state.
To Be Published
|
|
8WIW
 
 | | Cryo-EM structure of the flagellar C ring in the CW state | | Descriptor: | Chemotaxis protein CheY, Flagellar M-ring protein, Flagellar motor switch protein FliG, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-09-25 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Cryo-EM structure of the flagellar C ring in the CW state
To Be Published
|
|
8WOE
 
 | | Cryo-EM structure of the intact flagellar motor-hook complex in the CW state | | Descriptor: | Chemotaxis protein CheY, Flagellar L-ring protein, Flagellar M-ring protein, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-10-07 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of the intact flagellar motor-hook complex in the CW state
To Be Published
|
|
6PQ2
 
 | |
8V1Q
 
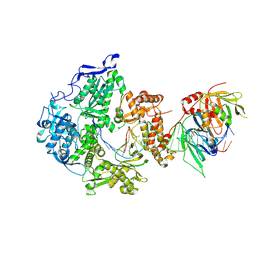 | | Herpes simplex virus 1 polymerase holoenzyme bound to DNA in both open/closed conformations | | Descriptor: | DNA polymerase, DNA polymerase processivity factor, MAGNESIUM ION, ... | | Authors: | Pan, J, Abraham, J, Coen, D.M, Shankar, S, Yang, P, Hogle, J. | | Deposit date: | 2023-11-21 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Viral DNA polymerase structures reveal mechanisms for drug selectivity
Cell(Cambridge,Mass.), 2024
|
|
8V1T
 
 | | Herpes simplex virus 1 polymerase holoenzyme bound to DNA and acyclovir triphosphate in closed conformation | | Descriptor: | ACYCLOVIR TRIPHOSPHATE, DNA polymerase, DNA polymerase processivity factor, ... | | Authors: | Pan, J, Abraham, J, Coen, D.M, Shankar, S, Yang, P, Hogle, J. | | Deposit date: | 2023-11-21 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Viral DNA polymerase structures reveal mechanisms for drug selectivity
Cell(Cambridge,Mass.), 2024
|
|
8WK4
 
 | |
8VPN
 
 | | Phosphorylated human NCC in complex with indapamide | | Descriptor: | 4-chloro-N-[(2S)-2-methyl-2,3-dihydro-1H-indol-1-yl]-3-sulfamoylbenzamide, ADENOSINE-5'-TRIPHOSPHATE, Solute carrier family 12 member 3 | | Authors: | Zhao, Y.X, Cao, E.H. | | Deposit date: | 2024-01-16 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural bases for Na + -Cl - cotransporter inhibition by thiazide diuretic drugs and activation by kinases.
Nat Commun, 15, 2024
|
|
6URS
 
 | | Sleeping Beauty transposase PAI subdomain mutant - H19Y | | Descriptor: | Sleeping Beauty transposase PAI subdomain | | Authors: | Nesmelova, I.V, Leighton, G.O, Yan, C, Lustig, J, Corona, R.I, Guo, J.T, Ivics, Z. | | Deposit date: | 2019-10-24 | | Release date: | 2020-10-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | H19Y mutation in the primary DNA-recognition subdomain of the Sleeping Beauty transposase improves structural stability, transposon DNA-binding and transposition
To Be Published
|
|
1B6Q
 
 | |
