2KBW
 
 | | Solution Structure of human Mcl-1 complexed with human Bid_BH3 peptide | | Descriptor: | BH3-interacting domain death agonist, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Liu, Q, Moldoveanu, T, Sprules, T, Matta-Camacho, E, Mansur-Azzam, N, Gehring, K. | | Deposit date: | 2008-12-09 | | Release date: | 2009-12-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Apoptotic regulation by MCL-1 through heterodimerization.
J.Biol.Chem., 285, 2010
|
|
2KTZ
 
 | | Inhibitor Induced Structural Change in the HCV IRES Domain IIa RNA | | Descriptor: | (7R)-7-[(dimethylamino)methyl]-1-[3-(dimethylamino)propyl]-7,8-dihydro-1H-furo[3,2-e]benzimidazol-2-amine, HCV IRES Domain IIa RNA | | Authors: | Paulsen, R.B, Seth, P.P, Swayze, E.E, Griffey, R.H, Skalicky, J.J, Cheatham III, T.E, Davis, D.R. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Inhibitor-induced structural change in the HCV IRES domain IIa RNA.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2KCU
 
 | | NMR solution structure of an uncharacterized protein from Chlorobium tepidum. Northeast Structural Genomics target CtR107 | | Descriptor: | protein CtR107 | | Authors: | Mills, J.L, Zhang, Q, Sukumaran, D.K, Wang, D, Jiang, M, Foote, E.L, Xiao, R, Nair, R, Everett, J.K, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-29 | | Release date: | 2009-01-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of an uncharacterized protein from Chlorobium tepidum. Northeast Structural Genomics target CtR107
To be Published
|
|
2KTU
 
 | |
2KPN
 
 | | Solution NMR structure of a Bacterial Ig-like (Big_3) domain from Bacillus cereus. Northeast Structural Genomics Consortium target BcR147A | | Descriptor: | Bacillolysin, CALCIUM ION | | Authors: | Aramini, J.M, Wang, D, Ciccosanti, C.T, Janjua, H, Rost, B, Acton, T.B, Xiao, R, Swapna, G.V.T, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-16 | | Release date: | 2010-01-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a Bacterial Ig-like (Big_3) domain from Bacillus cereus. Northeast Structural Genomics Consortium target BcR147A
To be Published
|
|
2M10
 
 | |
2MJ4
 
 | |
2MF9
 
 | | Solution structure of the N-terminal domain of human FKBP38 (FKBP38NTD) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP8 | | Authors: | Kang, C, Ye, H, Simon, B, Sattler, M, Yoon, H.S. | | Deposit date: | 2013-10-08 | | Release date: | 2013-11-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Functional role of the flexible N-terminal extension of FKBP38 in catalysis.
Sci Rep, 3, 2013
|
|
2MKI
 
 | | Solution structure of tandem RRM domains of cytoplasmic polyadenylation element binding protein 4 (CPEB4) in complex with RNA | | Descriptor: | Cytoplasmic polyadenylation element-binding protein 4, RNA (5'-R(*CP*UP*UP*UP*A)-3') | | Authors: | Afroz, T, Skrisovska, L, Belloc, E, Boixet, J.G, Mendez, R, Allain, F.H.-T. | | Deposit date: | 2014-02-07 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A fly trap mechanism provides sequence-specific RNA recognition by CPEB proteins
Genes Dev., 28, 2014
|
|
2MI0
 
 | | NMR structure of the I-V kissing-loop interaction of the Neurospora VS ribozyme | | Descriptor: | 5'-R(*GP*AP*GP*CP*AP*GP*CP*AP*UP*CP*GP*UP*CP*GP*GP*CP*UP*GP*CP*UP*CP*A)-3', 5'-R(*GP*CP*GP*GP*CP*AP*GP*UP*UP*GP*AP*CP*UP*AP*CP*UP*GP*UP*CP*GP*C)-3' | | Authors: | Bouchard, P, Legault, P. | | Deposit date: | 2013-12-05 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insights into substrate recognition by the neurospora varkud satellite ribozyme: importance of u-turns at the kissing-loop junction.
Biochemistry, 53, 2014
|
|
2MHQ
 
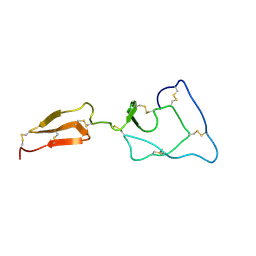 | | Solution structure of the major factor VIII binding region on von Willebrand factor | | Descriptor: | von Willebrand factor | | Authors: | Shiltagh, N, Kirkpatrick, J, Cabrita, L.D, McKinnon, T.A.J, Thalassinos, K, Tuddenham, E.G.D, Hansen, D.F. | | Deposit date: | 2013-12-02 | | Release date: | 2014-05-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the major factor VIII binding region on von Willebrand factor.
Blood, 123, 2014
|
|
2M6X
 
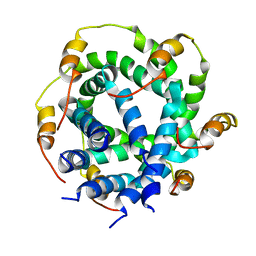 | |
2L16
 
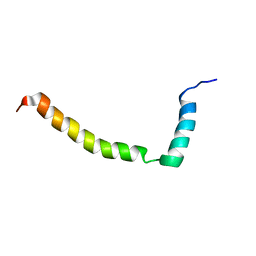 | |
2L28
 
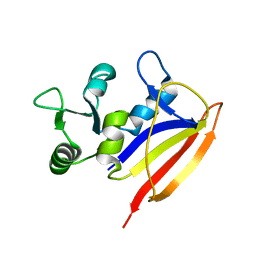 | | Solution structure of lactobacillus casei dihydrofolate reductase apo-form, 25 conformers | | Descriptor: | Dihydrofolate reductase | | Authors: | Polshakov, V.I, Birdsall, B, Feeney, J. | | Deposit date: | 2010-08-13 | | Release date: | 2011-04-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of Apo L. casei Dihydrofolate Reductase and Its Complexes with Trimethoprim and NADPH: Contributions to Positive Cooperative Binding from Ligand-Induced Refolding, Conformational Changes, and Interligand Hydrophobic Interactions.
Biochemistry, 50, 2011
|
|
2LA4
 
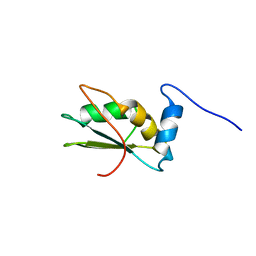 | | NMR structure of the C-terminal RRM domain of poly(U) binding 1 | | Descriptor: | Nuclear and cytoplasmic polyadenylated RNA-binding protein PUB1 | | Authors: | Santiveri, C.M, Mirassou, Y, Rico-Lastres, P, Martinez-Lumbreras, S, Perez-Canadillas, J.M. | | Deposit date: | 2011-03-01 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Pub1p C-terminal RRM domain interacts with Tif4631p through a conserved region neighbouring the Pab1p binding site
Plos One, 6, 2011
|
|
2LCV
 
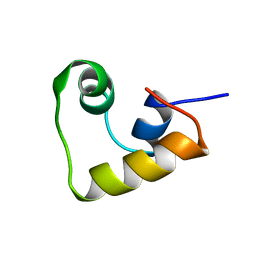 | |
2LGJ
 
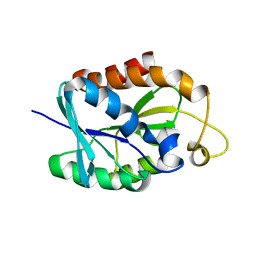 | | Solution structure of MsPTH | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Yadav, R, Pathak, P, Pulavarti, S, Jain, A, Kumar, A, Shukla, V, Arora, A. | | Deposit date: | 2011-07-27 | | Release date: | 2012-08-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of Peptidyl t-RNA hydrolase from Mycobacterium smegmatis
To be Published
|
|
2LQC
 
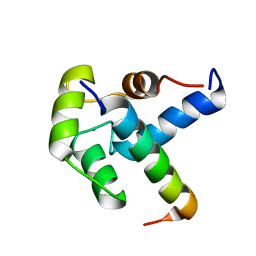 | |
2L7A
 
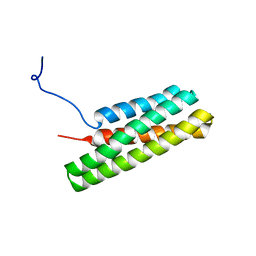 | | Solution Structure of the R3 Domain of Talin | | Descriptor: | Talin-1 | | Authors: | Goult, B.T, Gingras, A.R, Bate, N, Roberts, G.C.K, Barsukov, I.L, Critchley, D.R. | | Deposit date: | 2010-12-06 | | Release date: | 2011-12-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | RIAM and vinculin binding to talin are mutually exclusive and regulate adhesion assembly and turnover.
J.Biol.Chem., 288, 2013
|
|
2MJB
 
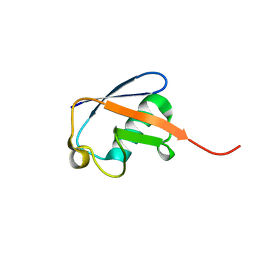 | | Solution nmr structure of ubiquitin refined against dipolar couplings in 4 media | | Descriptor: | Ubiquitin-60S ribosomal protein L40 | | Authors: | Maltsev, A, Grishaev, A, Roche, J, Zasloff, M, Bax, A. | | Deposit date: | 2014-01-02 | | Release date: | 2014-03-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Improved cross validation of a static ubiquitin structure derived from high precision residual dipolar couplings measured in a drug-based liquid crystalline phase.
J.Am.Chem.Soc., 136, 2014
|
|
2MJH
 
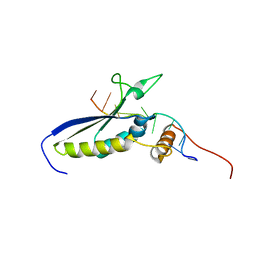 | |
2MA6
 
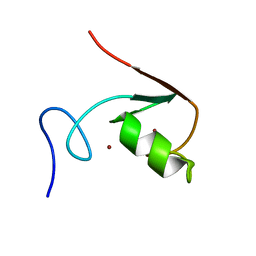 | | Solution NMR Structure of the RING finger domain from the Kip1 ubiquitination-promoting E3 complex protein 1 (KPC1/RNF123) from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR8700A | | Descriptor: | E3 ubiquitin-protein ligase RNF123, ZINC ION | | Authors: | Ramelot, T.A, Yang, Y, Janjua, H, Kohan, E, Wang, H, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-06-28 | | Release date: | 2013-07-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the RING finger domain from the Kip1 ubiquitination-promoting E3 complex protein 1 (KPC1/RNF123) from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR8700A
To be Published
|
|
2MKK
 
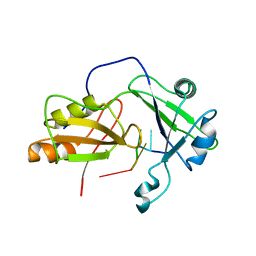 | | Structural model of tandem RRM domains of cytoplasmic polyadenylation element binding protein 1 (CPEB1) in complex with RNA | | Descriptor: | Cytoplasmic polyadenylation element-binding protein 1, RNA (5'-R(*UP*UP*UP*UP*A)-3') | | Authors: | Afroz, T, Skrisovska, L, Belloc, E, Boixet, J.G, Mendez, R, Allain, F.H.-T. | | Deposit date: | 2014-02-07 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A fly trap mechanism provides sequence-specific RNA recognition by CPEB proteins
Genes Dev., 28, 2014
|
|
2MLQ
 
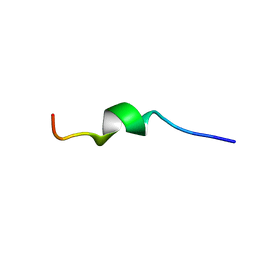 | | Human CCR2 Membrane-Proximal C-Terminal Region (PRO-C) in a frount bound form | | Descriptor: | MCP-1 receptor | | Authors: | Esaki, K, Yoshinaga, S, Tsuji, T, Toda, E, Terashima, Y, Saitoh, T, Kohda, D, Kohno, T, Osawa, M, Ueda, T, Shimada, I, Matsushima, K, Terasawa, H. | | Deposit date: | 2014-03-04 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the binding of the membrane-proximal C-terminal region of chemokine receptor CCR2 with the cytosolic regulator FROUNT.
Febs J., 281, 2014
|
|
2KI8
 
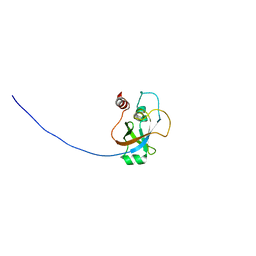 | | Solution NMR structure of tungsten formylmethanofuran dehydrogenase subunit D from Archaeoglobus fulgidus, Northeast Structural Genomics Consortium target AtT7 | | Descriptor: | Tungsten formylmethanofuran dehydrogenase, subunit D (FwdD-2) | | Authors: | Eletsky, A, Wu, Y, Yee, A, Fares, C, Lee, H.W, Arrowsmith, C.H, Prestegard, J.H, Szyperski, T, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-04-28 | | Release date: | 2009-05-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of tungsten formylmethanofuran dehydrogenase subunit D from Archaeoglobus fulgidus
To be Published
|
|
