4OC3
 
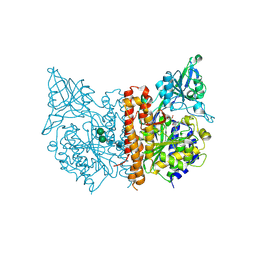 | | X-ray structure of of human glutamate carboxypeptidase II (GCPII) in a complex with CFIBzL, a urea-based inhibitor N~2~-{[(1S)-1-carboxy-2-(furan-2-yl)ethyl]carbamoyl}-N~6~-(4-iodobenzoyl)-L-lysine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pavlicek, J, Ptacek, J, Cerny, J, Byun, Y, Skultetyova, L, Pomper, M, Lubkowski, J, Barinka, C. | | Deposit date: | 2014-01-08 | | Release date: | 2014-05-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural characterization of P1'-diversified urea-based inhibitors of glutamate carboxypeptidase II.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4OCK
 
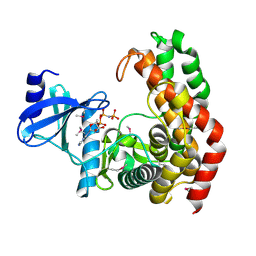 | | N-acetylhexosamine 1-phosphate kinase in complex with GlcNAc and AMPPNP | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, N-acetylhexosamine 1-phosphate kinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Li, T.L, Wang, K.C, Lyu, S.Y, Liu, Y.C, Chang, C.Y, Wu, C.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OEZ
 
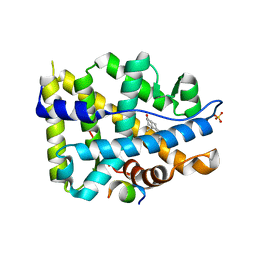 | | Crystal structure of AR-LBD bound with co-regulator peptide | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, SULFATE ION, ... | | Authors: | Liu, J.S, Hsu, C.L, Wu, W.G. | | Deposit date: | 2014-01-14 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
3V3S
 
 | | Crystal structure of GES-18 | | Descriptor: | Extended spectrum beta-lactamase GES-18 | | Authors: | Delbruck, H, Hoffmann, K.M.V, Bebrone, C. | | Deposit date: | 2011-12-14 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | GES-18, a new carbapenem-hydrolyzing GES-Type beta-lactamase from pseudomonas aeruginosa that contains Ile80 and Ser170 residues.
Antimicrob.Agents Chemother., 57, 2013
|
|
3V7A
 
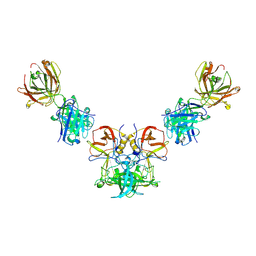 | |
4OF7
 
 | | Crystal Structure of SYG-1 D1, Crystal Form 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein SYG-1, isoform b, ... | | Authors: | Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-01-14 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
3V4A
 
 | | Structure of ar lbd with activator peptide and sarm inhibitor 2 | | Descriptor: | (5R)-3-(3,4-dichlorophenyl)-5-(4-hydroxyphenyl)-1,5-dimethyl-2-thioxoimidazolidin-4-one, Androgen receptor, SULFATE ION | | Authors: | Nique, F, Hebbe, S, Peixoto, C, Annoot, D, Lefrancois, J.-M, Duval, E, Michoux, L, Triballeau, N, Lemoullec, J.M, Mollat, P, Thauvin, M, Prange, T, Minet, D, Clement-Lacroix, P, Robin-Jagerschmidt, C, Fleury, D, Guedin, D, Deprez, P. | | Deposit date: | 2011-12-14 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of diarylhydantoins as new selective androgen receptor modulators.
J.Med.Chem., 55, 2012
|
|
4OD3
 
 | | Crystal structure of human Fab CAP256-VRC26.07, a potent V1V2-directed HIV-1 neutralizing antibody | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CAP256-VRC26.07 heavy chain, CAP256-VRC26.07 light chain, ... | | Authors: | Gorman, J, Doria-Rose, N.A, Schramm, C.A, Moore, P.L, Mascola, J.R, Shapiro, L, Morris, L, Kwong, P.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.616 Å) | | Cite: | Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies.
Nature, 509, 2014
|
|
3UIU
 
 | | Crystal structure of Apo-PKR kinase domain | | Descriptor: | Interferon-induced, double-stranded RNA-activated protein kinase | | Authors: | Li, F, Li, S, Yang, X, Shen, Y, Zhang, T. | | Deposit date: | 2011-11-06 | | Release date: | 2012-11-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Crystal structure of Apo-PKR kinase domain
TO BE PUBLISHED
|
|
4ODK
 
 | |
4OG2
 
 | | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with leucine | | Descriptor: | Amino acid/amide ABC transporter substrate-binding protein, HAAT family, CHLORIDE ION, ... | | Authors: | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-01-15 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.099 Å) | | Cite: | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with leucine
To be Published
|
|
3UJ7
 
 | | Phosphoethanolamine methyltransferase from Plasmodium falciparum in complex with SAM and PO4 | | Descriptor: | PHOSPHATE ION, Phosphoethanolamine N-methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Lee, S.G, Kim, Y, Alpert, T.D, Nagata, A, Jez, J.M. | | Deposit date: | 2011-11-07 | | Release date: | 2011-11-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Structure and reaction mechanism of phosphoethanolamine methyltransferase from the malaria parasite Plasmodium falciparum: an antiparasitic drug target.
J.Biol.Chem., 287, 2012
|
|
4ODR
 
 | | Structure of SlyD delta-IF from Thermus thermophilus in complex with FK506 | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, ACETATE ION, CHLORIDE ION, ... | | Authors: | Quistgaard, E.M, Low, C, Nordlund, P. | | Deposit date: | 2014-01-10 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.929 Å) | | Cite: | Molecular insights into substrate recognition and catalytic mechanism of the chaperone and FKBP peptidyl-prolyl isomerase SlyD.
BMC Biol., 14, 2016
|
|
2R9O
 
 | | Cathepsin S complexed with Compound 8 | | Descriptor: | Cathepsin S, N-[(1S)-2-{[(1R)-2-(benzyloxy)-1-cyano-1-methylethyl]amino}-1-(cyclohexylmethyl)-2-oxoethyl]morpholine-4-carboxamide | | Authors: | Ward, Y.D, Emmanuel, M.J, Thomson, D.S, Liu, W, Bekkali, Y, Frye, L.L, Girardot, M, Morwick, T, Young, E.R.R, Zindell, R, Hrapchak, M, DeTuri, M, White, A, Crane, K.M, White, D.M, Wang, Y, Hao, M.-H, Grygon, C.A, Labadia, M.E, Wildeson, J, Freeman, D, Nelson, R, Capolino, A, Peterson, J.D, Raymond, E.L, Brown, M.L, Spero, D.M. | | Deposit date: | 2007-09-13 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Synthesis of Reversible Inhibitors of Cathepsin S: alpha,alpha-Disubstitution at the P1 Residue Provides Potent Inhibitors in Cellular Assays and In Vivo Models of Antigen Presentation
to be published
|
|
4OGP
 
 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator (Space group : P21) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA topoisomerase 2-associated protein PAT1 | | Authors: | Fourati-Kammoun, Z, Kolesnikova, O, Back, R, Keller, J, Lazar, N, Gaudon-Plesse, C, Seraphin, B, Graille, M. | | Deposit date: | 2014-01-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The C-terminal domain from S. cerevisiae Pat1 displays two conserved regions involved in decapping factor recruitment.
Plos One, 9, 2014
|
|
3V4T
 
 | | E. cloacae C115D MURA liganded with UNAG | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, ... | | Authors: | Zhu, J.-Y, Yang, Y, Schonbrunn, E. | | Deposit date: | 2011-12-15 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Functional Consequence of Covalent Reaction of Phosphoenolpyruvate with UDP-N-acetylglucosamine 1-Carboxyvinyltransferase (MurA).
J.Biol.Chem., 287, 2012
|
|
4OH0
 
 | | Crystal structure of OXA-58 carbapenemase | | Descriptor: | Beta-lactamase OXA-58, CHLORIDE ION | | Authors: | Smith, C.A, Antunes, N.T, Toth, M, Vakulenko, S.B. | | Deposit date: | 2014-01-16 | | Release date: | 2014-02-26 | | Last modified: | 2014-04-02 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Carbapenemase OXA-58 from Acinetobacter baumannii.
Antimicrob.Agents Chemother., 58, 2014
|
|
3UKB
 
 | | (R)-cEt-BNA decamer structure | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(RCE)P*AP*CP*GP*C)-3') | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2011-11-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure and nuclease resistance of 2',4'-constrained 2'-O-methoxyethyl (cMOE) and 2'-O-ethyl (cEt) modified DNAs.
Chem.Commun.(Camb.), 48, 2012
|
|
4OES
 
 | | Crystal structure of NikA from Brucella suis in complex with Fe(III)-EDTA | | Descriptor: | FE (III) ION, GLYCEROL, NikA protein, ... | | Authors: | Lebrette, H, Cavazza, C. | | Deposit date: | 2014-01-13 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Promiscuous nickel import in human pathogens: structure, thermodynamics, and evolution of extracytoplasmic nickel-binding proteins.
Structure, 22, 2014
|
|
3ULN
 
 | |
4OHC
 
 | |
2RAY
 
 | |
3V5S
 
 | | Structure of Sodium/Calcium Exchanger from Methanococcus jannaschii | | Descriptor: | CADMIUM ION, Sodium/Calcium Exchanger | | Authors: | Liao, J, Li, H, Zeng, W, Sauer, D.B, Belmares, R, Jiang, Y. | | Deposit date: | 2011-12-16 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural insight into the ion-exchange mechanism of the sodium/calcium exchanger.
Science, 335, 2012
|
|
4OF9
 
 | | Structure of K42N variant of sperm whale myoglobin | | Descriptor: | 1,2-ETHANEDIOL, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lebioda, L, Wang, C, Lovelace, L.L. | | Deposit date: | 2014-01-14 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.241 Å) | | Cite: | Structures of K42N and K42Y sperm whale myoglobins point to an inhibitory role of distal water in peroxidase activity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OI8
 
 | | RAGE is a nucleic acid receptor that promotes inflammatory responses to DNA. | | Descriptor: | 5'-D(*CP*CP*AP*TP*GP*AP*CP*TP*GP*TP*AP*GP*GP*AP*AP*AP*CP*TP*CP*TP*AP*GP*A)-3', 5'-D(*CP*TP*CP*TP*AP*GP*AP*GP*TP*TP*TP*CP*CP*TP*AP*CP*AP*GP*TP*CP*AP*TP*G)-3', Advanced glycosylation end product-specific receptor | | Authors: | Jin, T, Jiang, J, Xiao, T. | | Deposit date: | 2014-01-19 | | Release date: | 2014-04-30 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | RAGE is a nucleic acid receptor that promotes inflammatory responses to DNA.
J.Exp.Med., 210, 2013
|
|
