4Y18
 
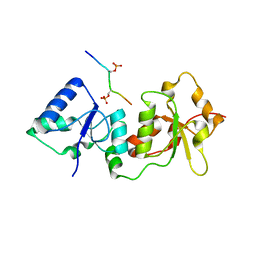 | |
4Y2G
 
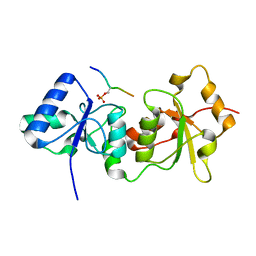 | |
8YR4
 
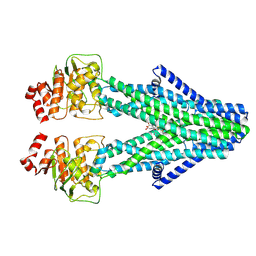 | | Cryo-EM structure of the human ABCB6 in complex with Cd(II):Phytochelatin 2 | | Descriptor: | ATP-binding cassette sub-family B member 6, CADMIUM ION, GAMMA-D-GLUTAMIC ACID, ... | | Authors: | Choi, S.H, Lee, S.S, Lee, H.Y, Kim, S, Kim, J.W, Jin, M.S. | | Deposit date: | 2024-03-20 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of cadmium-bound human ABCB6.
Commun Biol, 7, 2024
|
|
8YR3
 
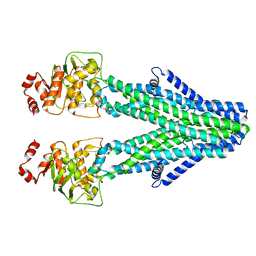 | | Cryo-EM structure of the human ABCB6 in complex with Cd(II):GSH | | Descriptor: | ATP-binding cassette sub-family B member 6, CADMIUM ION, GLUTATHIONE | | Authors: | Choi, S.H, Lee, S.S, Lee, H.Y, Kim, S, Kim, J.W, Jin, M.S. | | Deposit date: | 2024-03-20 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of cadmium-bound human ABCB6.
Commun Biol, 7, 2024
|
|
4EAQ
 
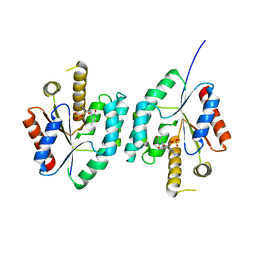 | | Crystal structure of Thymidylate Kinase from Staphylococcus aureus in complex with 3'-Azido-3'-Deoxythymidine-5'-Monophosphate | | Descriptor: | 3'-AZIDO-3'-DEOXYTHYMIDINE-5'-MONOPHOSPHATE, Thymidylate kinase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-03-22 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Thymidylate Kinase from Staphylococcus aureus in complex with 3'-Azido-3'-Deoxythymidine-5'-Monophosphate
To be Published
|
|
5KKK
 
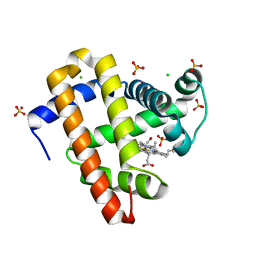 | |
4RQI
 
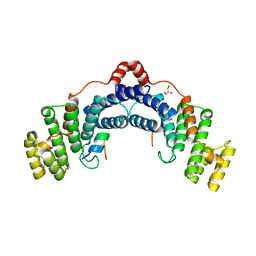 | | Structure of TRF2/RAP1 secondary interaction binding site | | Descriptor: | GLYCEROL, MAGNESIUM ION, Telomeric repeat-binding factor 2, ... | | Authors: | Miron, S, Guimaraes, B, Gaullier, G, Giraud-Panis, M.-J, Gilson, E, Le Du, M.-H. | | Deposit date: | 2014-11-03 | | Release date: | 2016-02-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4405 Å) | | Cite: | A higher-order entity formed by the flexible assembly of RAP1 with TRF2.
Nucleic Acids Res., 44, 2016
|
|
5HPD
 
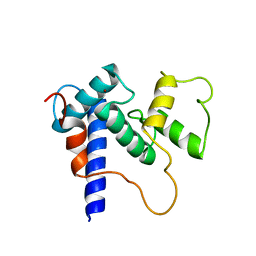 | | Solution Structure of TAZ2-p53TAD | | Descriptor: | CREB-binding protein,Cellular tumor antigen p53 fusion protein, ZINC ION | | Authors: | Krois, A.S, Ferreon, J.C, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2016-01-20 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of the disordered p53 transactivation domain by the transcriptional adapter zinc finger domains of CREB-binding protein.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5HOU
 
 | | Solution Structure of p53TAD-TAZ1 | | Descriptor: | Cellular tumor antigen p53,CREB-binding protein fusion protein, ZINC ION | | Authors: | Krois, A.S, Ferreon, J.C, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2016-01-19 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of the disordered p53 transactivation domain by the transcriptional adapter zinc finger domains of CREB-binding protein.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7Q33
 
 | |
1IH9
 
 | | NMR Structure of Zervamicin IIB (peptaibol antibiotic) Bound to DPC Micelles | | Descriptor: | ZERVAMICIN IIB | | Authors: | Shenkarev, Z.O, Balasheva, T.A, Efremov, R.G, Yakimenko, Z.A, Ovchinnikova, T.V, Raap, J, Arseniev, A.S. | | Deposit date: | 2001-04-19 | | Release date: | 2002-02-13 | | Last modified: | 2012-12-12 | | Method: | SOLUTION NMR | | Cite: | Spatial Structure of Zervamicin Iib Bound to Dpc Micelles: Implications for Voltage-Gating.
Biophys.J., 82, 2002
|
|
6Z1C
 
 | | Crystal structure of Arabidopsis thaliana CK2-alpha-1 in complex with TTP-22 | | Descriptor: | 3-[5-(4-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]sulfanylpropanoic acid, CHLORIDE ION, Casein kinase II subunit alpha-1 | | Authors: | Demulder, M, Loris, R, De Veylder, L. | | Deposit date: | 2020-05-13 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Arabidopsis casein kinase 2 triggers stem cell exhaustion under Al toxicity and phosphate deficiency through activating the DNA damage response pathway.
Plant Cell, 33, 2021
|
|
1U2A
 
 | |
1GXV
 
 | |
1GXX
 
 | |
1SSF
 
 | | Solution structure of the mouse 53BP1 fragment (residues 1463-1617) | | Descriptor: | Transformation related protein 53 binding protein 1 | | Authors: | Charier, G, Couprie, J, Alpha-Bazin, B, Meyer, V, Quemeneur, E, Guerois, R, Callebaut, I, Gilquin, B, Zinn-Justin, S. | | Deposit date: | 2004-03-24 | | Release date: | 2004-09-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Tudor Tandem of 53BP1; A New Structural Motif Involved in DNA and RG-Rich Peptide Binding
Structure, 12, 2004
|
|
8F2I
 
 | | P53 monomer structure | | Descriptor: | Cellular tumor antigen p53 | | Authors: | Solares, M, Kelly, D.F. | | Deposit date: | 2022-11-08 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | High-Resolution Imaging of Human Cancer Proteins Using Microprocessor Materials.
Chembiochem, 23, 2022
|
|
8F2H
 
 | |
7SGL
 
 | | DNA-PK complex of DNA end processing | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA-dependent protein kinase catalytic subunit, Hairpin_1, ... | | Authors: | Liu, L, Li, J, Chen, X, Yang, W, Gellert, M. | | Deposit date: | 2021-10-06 | | Release date: | 2022-01-12 | | Last modified: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Autophosphorylation transforms DNA-PK from protecting to processing DNA ends.
Mol.Cell, 82, 2022
|
|
2W0J
 
 | | Crystal structure of Chk2 in complex with NSC 109555, a specific inhibitor | | Descriptor: | 4,4'-DIACETYLDIPHENYLUREA-BIS(GUANYLHYDRAZONE), NITRATE ION, SERINE/THREONINE-PROTEIN KINASE CHK2 | | Authors: | Lountos, G.T, Tropea, J.E, Zhang, D, Jobson, A.G, Pommier, Y, Shoemaker, R.H, Waugh, D.S. | | Deposit date: | 2008-08-18 | | Release date: | 2009-02-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of Checkpoint Kinase 2 in Complex with Nsc 109555, a Potent and Selective Inhibitor
Protein Sci., 18, 2009
|
|
3MID
 
 | | Oxidized (Cu2+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound azide obtained by soaking (100mM NaN3) | | Descriptor: | AZIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-10 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Differential Reactivity Between the Two Copper Sites of Peptidylglycine alpha-Hydroxylating Monooxygenase (PHM)
J.Am.Chem.Soc., 132, 2010
|
|
3O4Z
 
 | |
3MIH
 
 | | Oxidized (Cu2+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound azide, obtained in the presence of substrate | | Descriptor: | AZIDE ION, COPPER (II) ION, IODIDE ION, ... | | Authors: | Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-10 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Differential Reactivity Between the Two Copper Sites of Peptidylglycine alpha-Hydroxylating Monooxygenase (PHM)
J.Am.Chem.Soc., 132, 2010
|
|
3MLK
 
 | | Reduced (Cu+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound nitrite | | Descriptor: | COPPER (II) ION, NICKEL (II) ION, NITRITE ION, ... | | Authors: | Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-16 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Differential reactivity between two copper sites in peptidylglycine alpha-hydroxylating monooxygenase
J.Am.Chem.Soc., 132, 2010
|
|
3MLJ
 
 | | Reduced (Cu+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound carbon monooxide (CO) | | Descriptor: | ACETATE ION, CARBON MONOXIDE, COPPER (II) ION, ... | | Authors: | Prigge, S.T, Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-16 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Differential reactivity between two copper sites in peptidylglycine alpha-hydroxylating monooxygenase
J.Am.Chem.Soc., 132, 2010
|
|
