6P1U
 
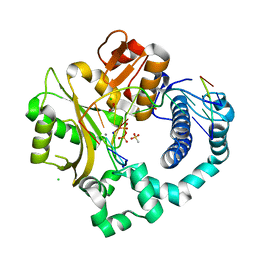 | | Post-catalytic nicked complex of human DNA Polymerase Mu with 1-nt gapped substrate containing template 8OG and newly incorporated CMP | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Kaminski, A.M, Pedersen, L.C, Bebenek, K, Chiruvella, K.K, Ramsden, D.A, Kunkel, T.A. | | Deposit date: | 2019-05-20 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Unexpected behavior of DNA polymerase Mu opposite template 8-oxo-7,8-dihydro-2'-guanosine.
Nucleic Acids Res., 47, 2019
|
|
8YQV
 
 | | African swine fever virus RNA Polymerase core | | Descriptor: | C122R, C147L, D339L, ... | | Authors: | Feng, X.Y. | | Deposit date: | 2024-03-20 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Regulation of early transcription and late transcript processing enzymes packaging for African swine fever virus from endogenous vRNAP-M1249L supercomplex structures
To Be Published
|
|
6C6S
 
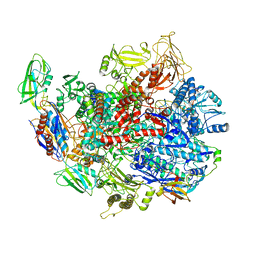 | | CryoEM structure of E.coli RNA polymerase elongation complex bound with RfaH | | Descriptor: | DNA (29-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Artsimovitch, I, Landick, R, Darst, S.A. | | Deposit date: | 2018-01-19 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis for Transcript Elongation Control by NusG Family Universal Regulators.
Cell, 173, 2018
|
|
6C6T
 
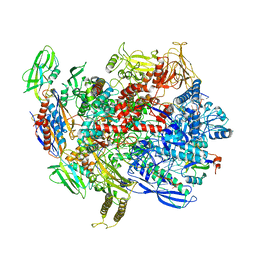 | | CryoEM structure of E.coli RNA polymerase elongation complex bound with RfaH | | Descriptor: | DNA (29-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Artsimovitch, I, Landick, R, Darst, S.A. | | Deposit date: | 2018-01-19 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Basis for Transcript Elongation Control by NusG Family Universal Regulators.
Cell, 173, 2018
|
|
6C6U
 
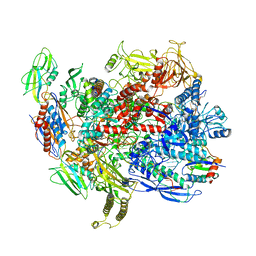 | | CryoEM structure of E.coli RNA polymerase elongation complex bound with NusG | | Descriptor: | DNA (29-MER), DNA-DIRECTED RNA POLYMERASE BETA', DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Kang, J.Y, Artsimovitch, I, Landick, R, Darst, S.A. | | Deposit date: | 2018-01-19 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis for Transcript Elongation Control by NusG Family Universal Regulators.
Cell, 173, 2018
|
|
4YCX
 
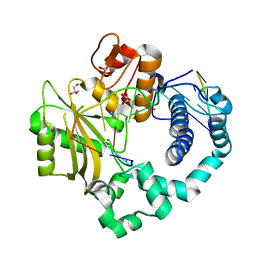 | | Binary complex of human DNA Polymerase Mu with 2-nt gapped DNA substrate | | Descriptor: | DNA (5'-D(*CP*GP*GP*CP*AP*AP*TP*AP*CP*G)-3'), DNA (5'-D(*CP*GP*TP*A)-3'), DNA (5'-D(P*GP*CP*CP*G)-3'), ... | | Authors: | Moon, A.F, Gosavi, R.A, Kunkel, T.A, Pedersen, L.C, Bebenek, K. | | Deposit date: | 2015-02-20 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Creative template-dependent synthesis by human polymerase mu.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8F5M
 
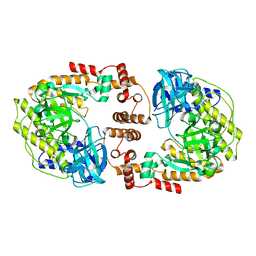 | | Crystal structure of P74 gp62 | | Descriptor: | Envelope glycoprotein gp62, MAGNESIUM ION | | Authors: | Bae, B, Nair, S.K. | | Deposit date: | 2022-11-14 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Tail-tape-fused virion and non-virion RNA polymerases of a thermophilic virus with an extremely long tail.
Nat Commun, 15, 2024
|
|
2XYM
 
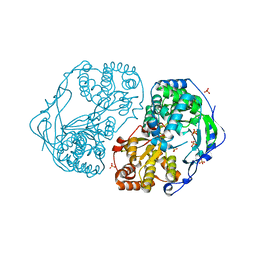 | | HCV-JFH1 NS5B T385A mutant | | Descriptor: | PHOSPHATE ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Simister, P.C, Caillet-Saguy, C, Bressanelli, S. | | Deposit date: | 2010-11-18 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.774 Å) | | Cite: | A Comprehensive Structure-Function Comparison of Hepatitis C Virus Strains Jfh1 and J6 Polymerases Reveals a Key Residue Stimulating Replication in Cell Culture Across Genotypes.
J.Virol., 85, 2011
|
|
4YD2
 
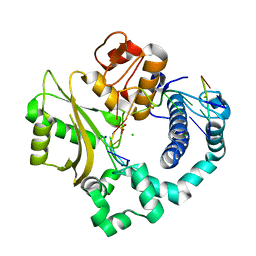 | | Nicked complex of human DNA Polymerase Mu with 2-nt gapped DNA substrate | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*GP*CP*AP*AP*T)-3'), DNA (5'-D(*CP*GP*TP*AP*T)-3'), ... | | Authors: | Moon, A.F, Gosavi, R.A, Kunkel, T.A, Pedersen, L.C, Bebenek, K. | | Deposit date: | 2015-02-20 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.471 Å) | | Cite: | Creative template-dependent synthesis by human polymerase mu.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6XEZ
 
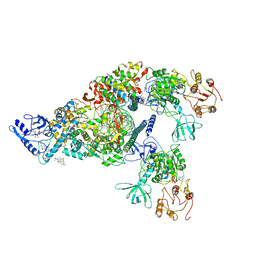 | | Structure of SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Llewellyn, E.C, Campbell, E.A, Darst, S.A. | | Deposit date: | 2020-06-14 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Basis for Helicase-Polymerase Coupling in the SARS-CoV-2 Replication-Transcription Complex.
Cell, 182, 2020
|
|
3CO9
 
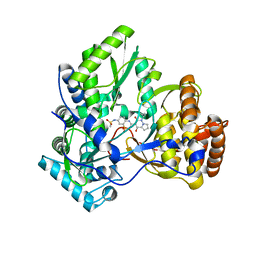 | | Crystal structure of HCV NS5B polymerase with a novel pyridazinone inhibitor | | Descriptor: | N-{3-[4-hydroxy-1-(3-methylbutyl)-2-oxo-1,2-dihydropyrrolo[1,2-b]pyridazin-3-yl]-1,1-dioxido-2H-1,2,4-benzothiadiazin-7 -yl}methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Han, Q, Showalter, R.E, Zhao, Q, Kissinger, C.R. | | Deposit date: | 2008-03-27 | | Release date: | 2009-02-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pyrrolo[1,2-b]pyridazin-2-ones as potent inhibitors of HCV NS5B polymerase.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
7UBK
 
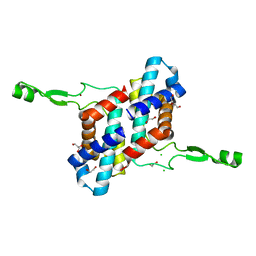 | | Transcription antitermination factor Qlambda, type-II crystal | | Descriptor: | 1,2-ETHANEDIOL, Antitermination protein Q, CHLORIDE ION, ... | | Authors: | Yin, Z, Ebright, R.H. | | Deposit date: | 2022-03-15 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | In transcription antitermination by Q lambda , NusA induces refolding of Q lambda to form a nozzle that extends the RNA polymerase RNA-exit channel.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UBJ
 
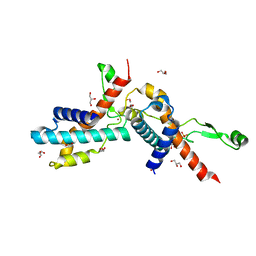 | | Transcription antitermination factor Qlambda, type-I crystal | | Descriptor: | 1,2-ETHANEDIOL, Antitermination protein Q, CHLORIDE ION, ... | | Authors: | Yin, Z, Ebright, R.H. | | Deposit date: | 2022-03-15 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | In transcription antitermination by Q lambda , NusA induces refolding of Q lambda to form a nozzle that extends the RNA polymerase RNA-exit channel.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UBL
 
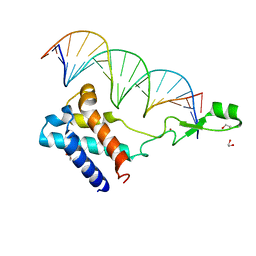 | | Transcription antitermination factor Qlambda in complex with Q-lambda-binding-element DNA | | Descriptor: | 1,2-ETHANEDIOL, Antitermination protein Q, DNA (5'-D(P*CP*AP*CP*CP*CP*AP*AP*TP*TP*TP*TP*AP*TP*TP*CP*AP*AP*TP*G)-3'), ... | | Authors: | Yin, Z, Ebright, R.H. | | Deposit date: | 2022-03-15 | | Release date: | 2022-09-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.177 Å) | | Cite: | In transcription antitermination by Q lambda , NusA induces refolding of Q lambda to form a nozzle that extends the RNA polymerase RNA-exit channel.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3SKA
 
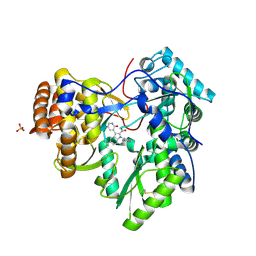 | |
7PLR
 
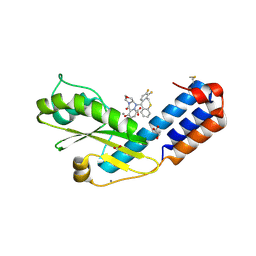 | | Crystal structure of the N-terminal endonuclease domain of La Crosse virus L-protein bound to compound Baloxavir | | Descriptor: | Baloxavir acid, FORMIC ACID, GLYCEROL, ... | | Authors: | Feracci, M, Hernandez, S, Vincentelli, R, Ferron, F, Reguera, J, Canard, B, Alvarez, K. | | Deposit date: | 2021-09-01 | | Release date: | 2022-09-14 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Biophysical and structural study of La Crosse virus endonuclease inhibition for the development of new antiviral options.
Iucrj, 11, 2024
|
|
4NYZ
 
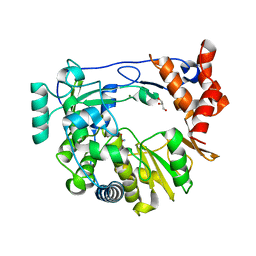 | | The EMCV 3Dpol structure with altered motif A conformation at 2.15A resolution | | Descriptor: | GLUTAMINE, GLYCEROL, Genome polyprotein, ... | | Authors: | Vives-adrian, L, Lujan, C, Ferrer-orta, C, Verdaguer, N. | | Deposit date: | 2013-12-11 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of a cardiovirus RNA-dependent RNA polymerase reveals an unusual conformation of the polymerase active site
J.Virol., 88, 2014
|
|
7CUN
 
 | | The structure of human Integrator-PP2A complex | | Descriptor: | Integrator complex subunit 1, Integrator complex subunit 11, Integrator complex subunit 2, ... | | Authors: | Zheng, H, Qi, Y, Liu, W, Li, J, Wang, J, Xu, Y. | | Deposit date: | 2020-08-23 | | Release date: | 2020-11-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Identification of Integrator-PP2A complex (INTAC), an RNA polymerase II phosphatase.
Science, 370, 2020
|
|
2BRK
 
 | |
2BRL
 
 | |
4NZ0
 
 | |
3D5M
 
 | | Crystal structure of HCV NS5B polymerase with a novel pyridazinone inhibitor | | Descriptor: | N-({3-[(5S)-5-tert-butyl-1-(3-chloro-4-fluorobenzyl)-4-hydroxy-2-oxo-2,5-dihydro-1H-pyrrol-3-yl]-1,1-dioxido-1,2-benzis othiazol-7-yl}methyl)methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Zhao, Q, Showalter, R.E, Han, Q, Kissinger, C.R. | | Deposit date: | 2008-05-16 | | Release date: | 2009-05-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based design, synthesis, and biological evaluation of 1,1-dioxoisothiazole and benzo[b]thiophene-1,1-dioxide derivatives as novel inhibitors of hepatitis C virus NS5B polymerase.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3CVK
 
 | | Crystal structure of HCV NS5B polymerase with a novel pyridazinone inhibitor | | Descriptor: | N-{3-[1-(3,3-Dimethyl-butyl)-4-hydroxy-2-oxo-1,2,4a,5,6,7-hexahydro-pyrrolo[1,2-b]pyridazin-3-yl]-1,1-dioxo-1,2-dihydro -1lambda6-benzo[1,2,4]thiadiazin-7-yl}-methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Zhao, Q, Showalter, R.E, Han, Q, Kissinger, C.R. | | Deposit date: | 2008-04-18 | | Release date: | 2009-04-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Hexahydro-pyrrolo- and hexahydro-1H-pyrido[1,2-b]pyridazin-2-ones as potent inhibitors of HCV NS5B polymerase.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
7DXY
 
 | |
7DXS
 
 | |
