5OOT
 
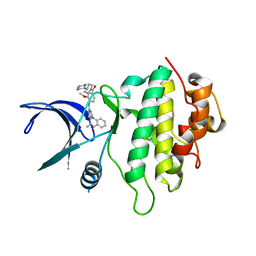 | | Structure of CHK1 10-pt. mutant complex with aminopyrimido-benzodiazepinone LRRK2 inhibitor | | Descriptor: | 2-[(2-methoxy-4-{[4-(4-methylpiperazin-1-yl)piperidin-1-yl]carbonyl}phenyl)amino]-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-08 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
5OWQ
 
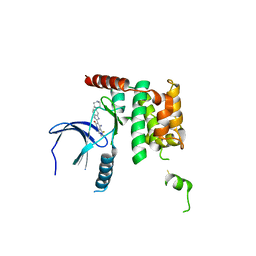 | | Human STK10 bound to dovitinib | | Descriptor: | 4-amino-5-fluoro-3-[5-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]quinolin-2(1H)-one, Serine/threonine-protein kinase 10 | | Authors: | Szklarz, M, von Delft, F, Bountra, C, Knapp, S, Edwards, A.M, Arrowsmith, C, Elkins, J.M. | | Deposit date: | 2017-09-04 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human STK10 bound to dovitinib
To Be Published
|
|
5OP2
 
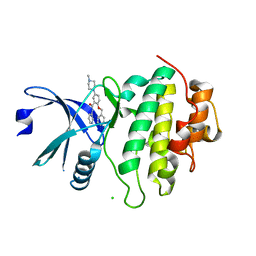 | | Structure of CHK1 10-pt. mutant complex with arylbenzamide LRRK2 inhibitor | | Descriptor: | 5-(4-methylpiperazin-1-yl)-2-phenylmethoxy-~{N}-pyridin-3-yl-benzamide, CHLORIDE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-09 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
3QUM
 
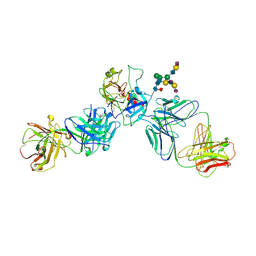 | | Crystal structure of human prostate specific antigen (PSA) in Fab sandwich with a high affinity and a PCa selective antibody | | Descriptor: | Fab 5D3D11 Heavy Chain, Fab 5D3D11 Light Chain, Fab 5D5A5 Heavy Chain, ... | | Authors: | Stura, E.A, Muller, B.H, Michel, S, Ducancel, F. | | Deposit date: | 2011-02-24 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of human prostate-specific antigen in a sandwich antibody complex.
J.Mol.Biol., 414, 2011
|
|
7VSY
 
 | | Pim1 with N82K mutation | | Descriptor: | Serine/threonine-protein kinase pim-1 | | Authors: | Hsu, C.Y, Tzeng, S.R. | | Deposit date: | 2021-10-27 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Pim1 with N82K mutation
To Be Published
|
|
5OXG
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with LDN-212854 | | Descriptor: | 1,2-ETHANEDIOL, 5-[6-(4-piperazin-1-ylphenyl)pyrazolo[1,5-a]pyrimidin-3-yl]quinoline, Activin receptor type-1, ... | | Authors: | Williams, E.P, Sorrell, F.J, Kopec, J, Nowak, R.P, Kupinska, K, von Delft, F, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-06 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural basis for the potent and selective binding of LDN-212854 to the BMP receptor kinase ALK2.
Bone, 109, 2018
|
|
7WBF
 
 | | Crystal structure of lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Nam, K.H. | | Deposit date: | 2021-12-16 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Processing of Multicrystal Diffraction Patterns in Macromolecular Crystallography Using Serial Crystallography Programs.
Crystals, 12, 2022
|
|
3S36
 
 | |
3S7L
 
 | |
7WBE
 
 | |
7WBD
 
 | |
3RVI
 
 | |
3RVR
 
 | | Structure of the CheYN59D/E89R Molybdate complex | | Descriptor: | Chemotaxis protein CheY, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Immormino, R.M, Starbird, C.A, Silversmith, R.E, Bourret, R.B. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing Mechanistic Similarities between Response Regulator Signaling Proteins and Haloacid Dehalogenase Phosphatases.
Biochemistry, 54, 2015
|
|
3RWO
 
 | | Crystal structure of YPT32 in complex with GDP | | Descriptor: | GTP-binding protein YPT32/YPT11, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Sultana, A, Jin, Y, Dregger, C, Franklin, E, Weisman, L.S, Khan, A.R. | | Deposit date: | 2011-05-09 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The activation cycle of Rab GTPase Ypt32 reveals structural determinants of effector recruitment and GDI binding.
Febs Lett., 585, 2011
|
|
3RXA
 
 | | Crystal structure of Trypsin complexed with cycloheptanamine | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXK
 
 | | Crystal structure of Trypsin complexed with methyl 4-amino-1-methyl-pyrrolidine-2-carboxylate | | Descriptor: | CALCIUM ION, Cationic trypsin, DIMETHYL SULFOXIDE, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXV
 
 | | Crystal structure of Trypsin complexed with benzamide (F05 and F03, cocktail experiment) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RVL
 
 | | Structure of the CheY-BeF3 Complex with substitutions at 59 and 89: N59D and E89R | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein CheY, GLYCEROL, ... | | Authors: | Starbird, C.A, Immormino, R.M, Silversmith, R.E, Bourret, R.B. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Probing Mechanistic Similarities between Response Regulator Signaling Proteins and Haloacid Dehalogenase Phosphatases.
Biochemistry, 54, 2015
|
|
7WKR
 
 | |
3S18
 
 | |
7W5O
 
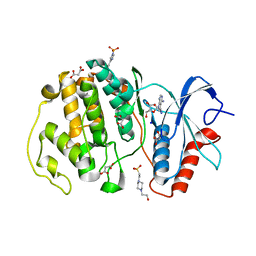 | | Crystal structure of ERK2 with an allosteric inhibitor | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 13-[4-({Imidazo[1,2-a]pyridin-2-yl}methoxy)phenyl]-4,8-dioxa-12,14,16,18-tetraazatetracyclo[9.7.0.0^{3,9}.0^{12,17}]octadeca-1(11),2,9,15,17-pentaen-15-amine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Yoshida, M, Kinoshita, T. | | Deposit date: | 2021-11-30 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification of a novel target site for ATP-independent ERK2 inhibitors.
Biochem.Biophys.Res.Commun., 593, 2022
|
|
3S8N
 
 | |
3S56
 
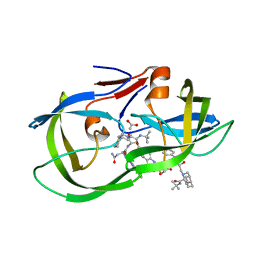 | | HIV-1 protease triple mutants V32I, I47V, V82I with antiviral drug saquinavir | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, ACETATE ION, Protease | | Authors: | Tie, Y.-F, Wang, Y.-F, Weber, I.T. | | Deposit date: | 2011-05-20 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Critical differences in HIV-1 and HIV-2 protease specificity for clinical inhibitors.
Protein Sci., 21, 2012
|
|
3S69
 
 | | Crystal structure of saxthrombin | | Descriptor: | CALCIUM ION, Thrombin-like enzyme defibrase | | Authors: | Huang, K, Zhao, W, Teng, M, Niu, L. | | Deposit date: | 2011-05-25 | | Release date: | 2012-05-23 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structure of saxthrombin, a thrombin-like enzyme from Gloydius saxatilis.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3RZ4
 
 | |
