4UY1
 
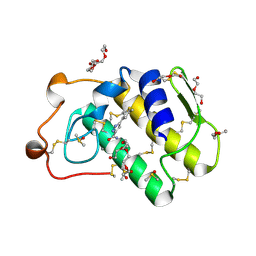 | | Novel pyrazole series of group X Secretory Phospholipase A2 (sPLA2-X) inhibitors | | Descriptor: | 5-(2,5-DIMETHYL-3-THIENYL)-1H-PYRAZOLE-3-CARBOXAMIDE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sandmark, J, Oster, L, Hallberg, K, Bodin, C, Chen, H. | | Deposit date: | 2014-08-28 | | Release date: | 2014-10-15 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a Novel Pyrazole Series of Group X Secreted Phospholipase A2 Inhibitor (Spla2X) Via Fragment Based Virtual Screening
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4URD
 
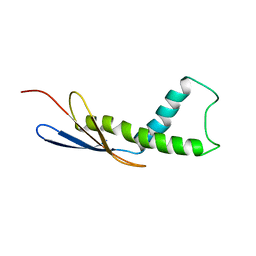 | | Cryo-EM map of Trigger Factor bound to a translating ribosome | | Descriptor: | TRIGGER FACTOR | | Authors: | Deeng, J, Chan, K.Y, van der Sluis, E, Bischoff, L, Berninghausen, O, Han, W, Gumbart, J, Schulten, K, Beatrix, B, Beckmann, R. | | Deposit date: | 2014-06-27 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Dynamic Behavior of Trigger Factor on the Ribosome.
J.Mol.Biol., 428, 2016
|
|
4UZ0
 
 | | Crystal Structure of apoptosis repressor with CARD (ARC) | | Descriptor: | GLYCEROL, NUCLEOLAR PROTEIN 3 | | Authors: | Kim, S.H, Jeong, J.H, Jang, T.H, Kim, Y.G, Park, H.H. | | Deposit date: | 2014-09-04 | | Release date: | 2015-07-01 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Crystal Structure of Caspase Recruiting Domain (Card) of Apoptosis Repressor with Card (Arc) and its Implication in Inhibition of Apoptosis.
Sci.Rep., 5, 2015
|
|
4V0S
 
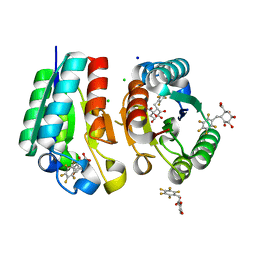 | | Crystal structure of Mycobacterium tuberculosis Type II Dehydroquinase D88N mutant inhibited by a 3-dehydroquinic acid derivative | | Descriptor: | (1R,2S,4S,5R)-2-(2,3,4,5,6-pentafluorophenyl)methyl-1,4,5-trihydroxy-3-oxocyclohexane-1-carboxylic acid, 3,4-DIHYDROXY-2-[(2,3,4,5,6-PENTAFLUOROPHENYL)METHYL]BENZOIC ACID, 3-DEHYDROQUINATE DEHYDRATASE, ... | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Santiago, C, Lamb, H, Hawkins, A.R, Maneiro, M, Peon, A, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2014-09-18 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Investigation of the Dehydratation Mechanism Catalyzed by the Type II Dehydroquinase
To be Published
|
|
4UZ9
 
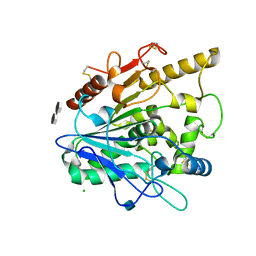 | | STRUCTURE OF THE WNT DEACYLASE NOTUM - CRYSTAL FORM VII - SOS COMPLEX - 2.2A | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Zebisch, M, Jones, E.Y. | | Deposit date: | 2014-09-04 | | Release date: | 2015-02-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Notum Deacylates Wnt Proteins to Suppress Signalling Activity.
Nature, 519, 2015
|
|
4UQO
 
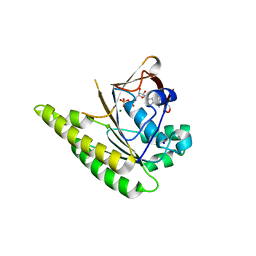 | | RADA C-TERMINAL ATPASE DOMAIN FROM PYROCOCCUS FURIOSUS BOUND TO ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA REPAIR AND RECOMBINATION PROTEIN RADA, MAGNESIUM ION, ... | | Authors: | Marsh, M.E, Ehebauer, M.T, Scott, D, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2014-06-24 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | ATP Half-Sites in Rada and Rad51 Recombinases Bind Nucleotides
FEBS Open Bio, 6, 2016
|
|
4UZL
 
 | |
4V0X
 
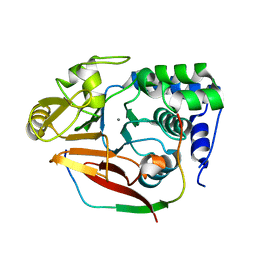 | | The crystal structure of mouse PP1G in complex with truncated human PPP1R15B (631-684) | | Descriptor: | MANGANESE (II) ION, PROTEIN PHOSPHATASE 1 REGULATORY SUBUNIT 15B, PROTEIN PHOSPHATASE PP1-GAMMA CATALYTIC SUBUNIT | | Authors: | Chen, R, Yan, Y, Casado, A.C, Ron, D, Read, R.J. | | Deposit date: | 2014-09-18 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | G-actin provides substrate-specificity to eukaryotic initiation factor 2 alpha holophosphatases.
Elife, 4, 2015
|
|
4USI
 
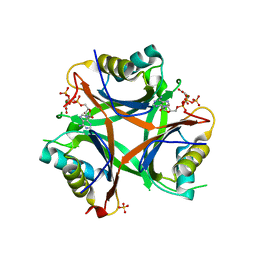 | | Nitrogen regulatory protein PII from Chlamydomonas reinhardtii in complex with MgATP and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Chellamuthu, V.R, Forchhammer, K, Hartmann, M.D. | | Deposit date: | 2014-07-08 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A Widespread Glutamine-Sensing Mechanism in the Plant Kingdom.
Cell(Cambridge,Mass.), 159, 2014
|
|
4V18
 
 | | SeMet structure of a novel carbohydrate binding module from glycoside hydrolase family 5 glucanase from Ruminococcus flavefaciens FD-1 | | Descriptor: | CARBOHYDRATE BINDING MODULE | | Authors: | Venditto, I, Centeno, M.S.J, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-25 | | Release date: | 2016-01-20 | | Last modified: | 2016-07-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Complexity of the Ruminococcus Flavefaciens Cellulosome Reflects an Expansion in Glycan Recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4USR
 
 | | Structure of flavin-containing monooxygenase from Pseudomonas stutzeri NF13 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, MONOOXYGENASE | | Authors: | Jensen, C.N, Ali, S.T, Allen, M.J, Grogan, G. | | Deposit date: | 2014-07-11 | | Release date: | 2014-10-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Exploring Nicotinamide Cofactor Promiscuity in Nad(P)H-Dependent Flavin Containing Monooxygenases (Fmos) Using Natural Variation within the Phosphate Binding Loop. Structure and Activity of Fmos from Cellvibrio Sp. Br and Pseudomonas Stutzeri NF13
J.Mol.Catal., 109, 2014
|
|
4V3F
 
 | |
4USX
 
 | | The Structure of the C-terminal YadA-like domain of BPSL2063 from Burkholderia pseudomallei | | Descriptor: | MAGNESIUM ION, TRIMERIC AUTOTRANSPORTER ADHESIN | | Authors: | Perletti, L, Gourlay, L.J, Peano, C, Pietrelli, A, DeBellis, G, Deantonio, C, Santoro, C, Sblattero, D, Bolognesi, M. | | Deposit date: | 2014-07-16 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Selecting Soluble/Foldable Protein Domains Through Single-Gene or Genomic Orf Filtering: Structure of the Head Domain of Burkholderia Pseudomallei Antigen Bpsl2063.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4V4D
 
 | | Crystal Structure of Pyrogallol-Phloroglucinol Transhydroxylase from Pelobacter acidigallici complexed with pyrogallol | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, BENZENE-1,2,3-TRIOL, CALCIUM ION, ... | | Authors: | Messerschmidt, A, Niessen, H, Abt, D, Einsle, O, Schink, B, Kroneck, P.M.H. | | Deposit date: | 2004-06-02 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of pyrogallol-phloroglucinol transhydroxylase, an Mo enzyme capable of intermolecular hydroxyl transfer between phenols
PROC.NATL.ACAD.SCI.USA, 101, 2004
|
|
4UTP
 
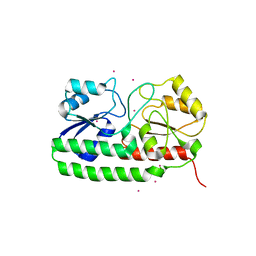 | | Crystal structure of pneumococcal surface antigen PsaA in the Cd- bound, closed state | | Descriptor: | CADMIUM ION, MANGANESE ABC TRANSPORTER SUBSTRATE-BINDING LIPOPROTEIN | | Authors: | Luo, Z, Counago, R.M, Maher, M, Kobe, B. | | Deposit date: | 2014-07-22 | | Release date: | 2014-08-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dysregulation of transition metal ion homeostasis is the molecular basis for cadmium toxicity in Streptococcus pneumoniae.
Nat Commun, 6, 2015
|
|
4V4N
 
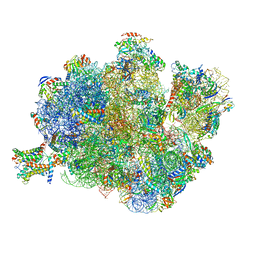 | | Structure of the Methanococcus jannaschii ribosome-SecYEBeta channel complex | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein L7AE, ... | | Authors: | Menetret, J.F, Park, E, Gumbart, J.C, Ludtke, S.J, Li, W, Whynot, A, Rapoport, T.A, Akey, C.W. | | Deposit date: | 2013-06-17 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structure of the SecY channel during initiation of protein translocation.
Nature, 506, 2013
|
|
4UV9
 
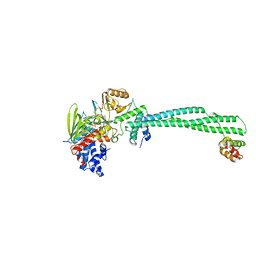 | | LSD1(KDM1A)-CoREST in complex with 1-Ethyl-Tranylcypromine | | Descriptor: | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S)-2,3,4-trihydroxy-5-[(4aS,10aS)-4a-[(1S,3E)-3-imino-1-phenylpentyl]-7,8-dimethyl-2,4-dioxo-1,3,4,4a,5,10a-hexahydrobenzo[g]pteridin-10(2H)-yl]pentyl dihydrogen diphosphate | | Authors: | Vianello, P, Botrugno, O, Cappa, A, Ciossani, G, Dessanti, P, Mai, A, Mattevi, A, Meroni, G, Minucci, S, Thaler, F, Tortorici, M, Trifiro, P, Valente, S, Villa, M, Varasi, M, Mercurio, C. | | Deposit date: | 2014-08-05 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis, Biological Activity and Mechanistic Insights of 1-Substituted Cyclopropylamine Derivatives: A Novel Class of Irreversible Inhibitors of Histone Demethylase Kdm1A.
Eur.J.Med.Chem., 86C, 2014
|
|
4UXL
 
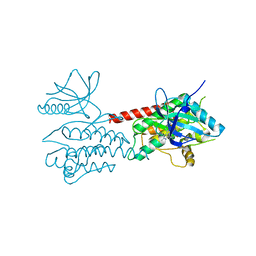 | | Structure of Human ROS1 Kinase Domain in Complex with PF-06463922 | | Descriptor: | (10R)-7-amino-12-fluoro-2,10,16-trimethyl-15-oxo-10,15,16,17-tetrahydro-2H-8,4-(metheno)pyrazolo[4,3-h][2,5,11]benzoxadiazacyclotetradecine-3-carbonitrile, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE ROS | | Authors: | McTigue, M, Deng, Y, Liu, W, Brooun, A, Stewart, A. | | Deposit date: | 2014-08-25 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Pf-06463922 is a Potent and Selective Next-Generation Ros1/Alk Inhibitor Capable of Blocking Crizotinib-Resistant Ros1 Mutations.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4UYQ
 
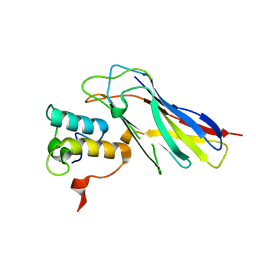 | |
4V1L
 
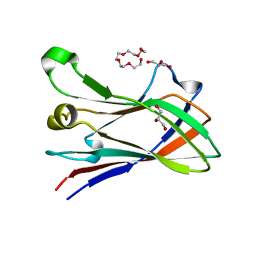 | | High resolution structure of a novel carbohydrate binding module from glycoside hydrolase family 9 (Cel9A) from Ruminococcus flavefaciens FD-1 | | Descriptor: | CARBOHYDRATE BINDING MODULE, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Venditto, I, Goyal, A, Thompson, A, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-29 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Complexity of the Ruminococcus Flavefaciens Cellulosome Reflects an Expansion in Glycan Recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4UUM
 
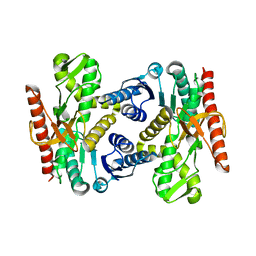 | |
4UX6
 
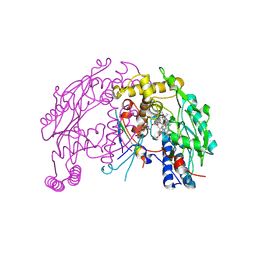 | | The discovery of novel, potent and highly selective inhibitors of inducible nitric oxide synthase (iNOS) | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, NITRIC OXIDE SYNTHASE, INDUCIBLE, ... | | Authors: | Cheshire, D.R, Andrews, G, Beaton, H.G, Birkinshaw, T, Boughton-Smith, N, Connolly, S, Cook, T.R, Cooper, A, Cooper, S.L, Cox, D, Dixon, J, Gensmantel, N, Hamley, P.J, Harrison, R, Hartopp, P, Kack, H, Luker, T, Mete, A, Millichip, I, Nicholls, D.J, Pimm, A.D, St-Gallay, S.A, Wallace, A.V. | | Deposit date: | 2014-08-19 | | Release date: | 2014-10-08 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Discovery of Novel, Potent and Highly Selective Inhibitors of Inducible Nitric Oxide Synthase (Inos).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
5HD1
 
 | | Crystal structure of antimicrobial peptide Pyrrhocoricin bound to the Thermus thermophilus 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Gagnon, M.G, Roy, R.N, Lomakin, I.B, Florin, T, Mankin, A.S, Steitz, T.A. | | Deposit date: | 2016-01-04 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of proline-rich peptides bound to the ribosome reveal a common mechanism of protein synthesis inhibition.
Nucleic Acids Res., 44, 2016
|
|
5J4C
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with cisplatin (soaked) and bound to mRNA and A-, P- and E-site tRNAs at 2.8A resolution | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Melnikov, S.V, Soll, D, Steitz, T.A, Polikanov, Y.S. | | Deposit date: | 2016-03-31 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insights into RNA binding by the anticancer drug cisplatin from the crystal structure of cisplatin-modified ribosome.
Nucleic Acids Res., 44, 2016
|
|
5HCP
 
 | | Crystal structure of antimicrobial peptide Metalnikowin bound to the Thermus thermophilus 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Gagnon, M.G, Roy, R.N, Lomakin, I.B, Florin, T, Mankin, A.S, Steitz, T.A. | | Deposit date: | 2016-01-04 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Structures of proline-rich peptides bound to the ribosome reveal a common mechanism of protein synthesis inhibition.
Nucleic Acids Res., 44, 2016
|
|
