5VLZ
 
 | | Backbone model for phage Qbeta capsid | | Descriptor: | Capsid protein, Maturation protein A2 | | Authors: | Cui, Z, Zhang, J. | | Deposit date: | 2017-04-26 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structures of Q beta virions, virus-like particles, and the Q beta-MurA complex reveal internal coat proteins and the mechanism of host lysis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2QUX
 
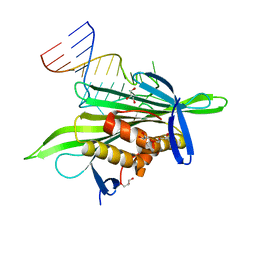 | | PP7 coat protein dimer in complex with RNA hairpin | | Descriptor: | Coat protein, GLYCEROL, RNA (25-MER) | | Authors: | Chao, J.A. | | Deposit date: | 2007-08-06 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural basis for the coevolution of a viral RNA-protein complex.
Nat.Struct.Mol.Biol., 15, 2008
|
|
6E5K
 
 | | Heterogeneous-Backbone Mimics of a Designed Disulfide-Rich Protein: Aib turn, Aib helix, N-methyl hairpin | | Descriptor: | Designed peptide NC_HEE_D1: Aib turn, Aib helix, N-methyl hairpin mutant | | Authors: | Cabalteja, C.C, Mihalko, D.S, Horne, W.S. | | Deposit date: | 2018-07-20 | | Release date: | 2018-11-21 | | Last modified: | 2020-01-01 | | Method: | SOLUTION NMR | | Cite: | Heterogeneous-Backbone Foldamer Mimics of a Computationally Designed, Disulfide-Rich Miniprotein.
Chembiochem, 20, 2019
|
|
6ZZR
 
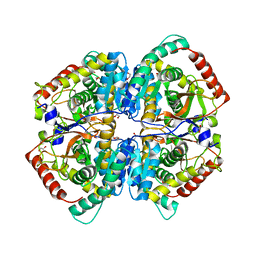 | | The Crystal Structure of human LDHA from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, L-lactate dehydrogenase A chain | | Authors: | Wang, F, Lin, D, Cheng, W, Bao, X, Zhu, B, Shang, H. | | Deposit date: | 2020-08-05 | | Release date: | 2020-08-19 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Crystal Structure of human LDHA from Biortus
To Be Published
|
|
1SL6
 
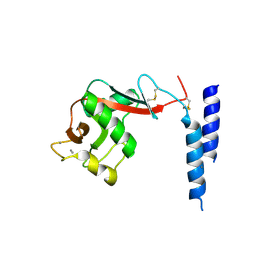 | | Crystal Structure of a fragment of DC-SIGNR (containg the carbohydrate recognition domain and two repeats of the neck) complexed with Lewis-x. | | Descriptor: | C-type lectin DC-SIGNR, CALCIUM ION, alpha-L-fucopyranose-(1-3)-[beta-D-galactopyranose-(1-4)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Guo, Y, Feinberg, H, Conroy, E, Mitchell, D.A, Alvarez, R, Blixt, O, Taylor, M.E, Weis, W.I, Drickamer, K. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for distinct ligand-binding and targeting properties of the receptors
DC-SIGN and DC-SIGNR
Nat.Struct.Mol.Biol., 11, 2004
|
|
2DGC
 
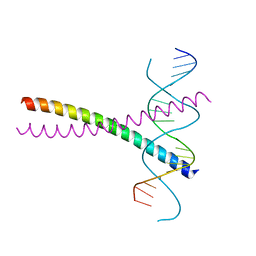 | |
6ZVR
 
 | | C11 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | Vipp1 | | Authors: | Liu, J.W, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6ZW4
 
 | | C14 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | vipp1 | | Authors: | Liu, J.W, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6ZW5
 
 | | C15 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | vipp1 | | Authors: | Liu, J.W, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6ZVS
 
 | | C12 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | Vipp1 | | Authors: | Liu, J, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6ZVT
 
 | | C13 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | Vipp1 | | Authors: | Liu, J.W, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
1J09
 
 | | Crystal structure of Thermus thermophilus glutamyl-tRNA synthetase complexed with ATP and Glu | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLUTAMIC ACID, Glutamyl-tRNA synthetase, ... | | Authors: | Sekine, S, Nureki, O, Dubois, D.Y, Bernier, S, Chenevert, R, Lapointe, J, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-11-12 | | Release date: | 2003-02-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | ATP binding by glutamyl-tRNA synthetase is switched to the productive mode by tRNA binding
EMBO J., 22, 2003
|
|
6ZW6
 
 | | C16 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | vipp1 | | Authors: | Liu, J, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6ZW7
 
 | | C17 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | vipp1 | | Authors: | Liu, J, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
1TNZ
 
 | | Rat Protein Geranylgeranyltransferase Type-I Complexed with a GGPP analog and a RRCVLL Peptide Derived from Cdc42 splice isoform-2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[METHYL-(5-GERANYL-4-METHYL-PENT-3-ENYL)-AMINO]-ETHYL-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Reid, T.S, Terry, K.L, Casey, P.J, Beese, L.S. | | Deposit date: | 2004-06-11 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic analysis of CaaX prenyltransferases complexed with substrates defines rules of protein substrate selectivity.
J.Mol.Biol., 343, 2004
|
|
2DER
 
 | | Cocrystal structure of an RNA sulfuration enzyme MnmA and tRNA-Glu in the initial tRNA binding state | | Descriptor: | PHOSPHATE ION, SULFATE ION, tRNA, ... | | Authors: | Numata, T, Ikeuchi, Y, Fukai, S, Suzuki, T, Nureki, O. | | Deposit date: | 2006-02-17 | | Release date: | 2006-08-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Snapshots of tRNA sulphuration via an adenylated intermediate
Nature, 442, 2006
|
|
6E8G
 
 | |
1S1L
 
 | |
1IHF
 
 | | INTEGRATION HOST FACTOR/DNA COMPLEX | | Descriptor: | CADMIUM ION, DNA (35-MER), DNA (5'-D(*GP*CP*TP*TP*AP*TP*CP*AP*AP*TP*TP*TP*GP*TP*TP*GP*C P*AP*CP*C)-3'), ... | | Authors: | Rice, P.A, Yang, S.-W, Mizuuchi, K, Nash, H.A. | | Deposit date: | 1996-08-21 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn.
Cell(Cambridge,Mass.), 87, 1996
|
|
2DET
 
 | | Cocrystal structure of an RNA sulfuration enzyme MnmA and tRNA-Glu in the pre-reaction state | | Descriptor: | SULFATE ION, tRNA, tRNA-specific 2-thiouridylase mnmA | | Authors: | Numata, T, Ikeuchi, Y, Fukai, S, Suzuki, T, Nureki, O. | | Deposit date: | 2006-02-17 | | Release date: | 2006-08-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Snapshots of tRNA sulphuration via an adenylated intermediate
Nature, 442, 2006
|
|
1SGH
 
 | | Moesin FERM domain bound to EBP50 C-terminal peptide | | Descriptor: | Ezrin-radixin-moesin binding phosphoprotein 50, Moesin | | Authors: | Finnerty, C.M, Chambers, D, Ingraffea, J, Faber, H.R, Karplus, P.A, Bretscher, A. | | Deposit date: | 2004-02-23 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The EBP50-moesin interaction involves a binding site regulated by direct masking on the FERM domain
J.Cell.Sci., 117, 2004
|
|
2BUA
 
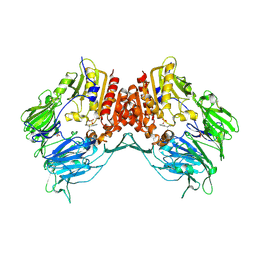 | | Crystal Structure Of Porcine Dipeptidyl Peptidase IV (Cd26) in Complex With a Low Molecular Weight Inhibitor. | | Descriptor: | 1-METHYLAMINE-1-BENZYL-CYCLOPENTANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nordhoff, S, Cerezo-Galvez, S, Feurer, A, Hill, O, Matassa, V.G, Metz, G, Rummey, C, Thiemann, M, Edwards, P.J. | | Deposit date: | 2005-06-09 | | Release date: | 2006-01-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The reversed binding of beta-phenethylamine inhibitors of DPP-IV: X-ray structures and properties of novel fragment and elaborated inhibitors.
Bioorg. Med. Chem. Lett., 16, 2006
|
|
1IJW
 
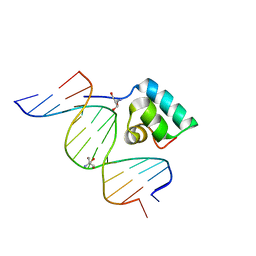 | | Testing the Water-Mediated Hin Recombinase DNA Recognition by Systematic Mutations. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*AP*TP*(CBR)P*TP*TP*AP*TP*CP*AP*AP*AP*AP*AP*C)-3', 5'-D(*TP*GP*TP*TP*TP*TP*TP*GP*AP*TP*AP*AP*GP*A)-3', ... | | Authors: | Chiu, T.K, Sohn, C, Johnson, R.C, Dickerson, R.E. | | Deposit date: | 2001-04-30 | | Release date: | 2002-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Testing water-mediated DNA recognition by the Hin recombinase.
EMBO J., 21, 2002
|
|
6E5H
 
 | |
1RCX
 
 | |
