6K7M
 
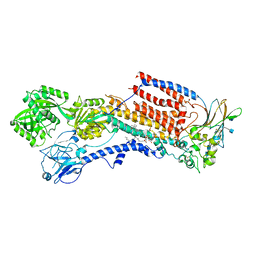 | | Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E2Pi-PL state) | | Descriptor: | (2~{S})-2-azanyl-3-[[(2~{R})-3-hexadecanoyloxy-2-[(~{Z})-octadec-9-enoyl]oxy-propoxy]-oxidanyl-phosphoryl]oxy-propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Hiraizumi, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-06-07 | | Release date: | 2019-08-28 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Cryo-EM structures capture the transport cycle of the P4-ATPase flippase.
Science, 365, 2019
|
|
7M5C
 
 | |
5VUS
 
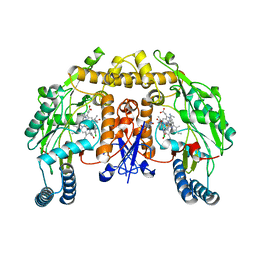 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 4-(2-(((2-Aminoquinolin-7-yl)methyl)amino)ethyl)-2-chlorobenzonitrile | | Descriptor: | 4-(2-{[(2-aminoquinolin-7-yl)methyl]amino}ethyl)-2-chlorobenzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Hydrophilic, Potent, and Selective 7-Substituted 2-Aminoquinolines as Improved Human Neuronal Nitric Oxide Synthase Inhibitors.
J. Med. Chem., 60, 2017
|
|
2OPW
 
 | | Crystal structure of human phytanoyl-CoA dioxygenase PHYHD1 (apo) | | Descriptor: | PHYHD1 protein | | Authors: | Zhang, Z, Butler, D, McDonough, M.A, Kavanagh, K.L, Bray, J.E, Ng, S.S, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-30 | | Release date: | 2007-03-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human phytanoyl-CoA dioxygenase PHYHD1 (apo)
To be Published
|
|
7MFX
 
 | |
5VV3
 
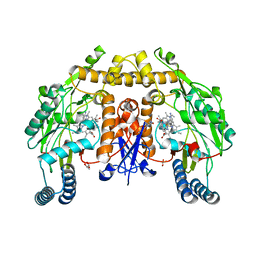 | | Structure of human neuronal nitric oxide synthase heme domain in complex with 4-(2-(((2-Aminoquinolin-7-yl)methyl)amino)ethyl)benzonitrile | | Descriptor: | 4-(2-{[(2-aminoquinolin-7-yl)methyl]amino}ethyl)benzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Hydrophilic, Potent, and Selective 7-Substituted 2-Aminoquinolines as Improved Human Neuronal Nitric Oxide Synthase Inhibitors.
J. Med. Chem., 60, 2017
|
|
5VJZ
 
 | | Joint X-ray/neutron structure of aspartate aminotransferase with alpha-methyl-aspartate at pH 7.5 | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-2-METHYL-SUCCINIC ACID, Aspartate aminotransferase, cytoplasmic | | Authors: | Dajnowicz, S, Kovalevsky, A.Y, Mueser, T.C. | | Deposit date: | 2017-04-20 | | Release date: | 2017-11-01 | | Last modified: | 2022-03-16 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | Direct visualization of critical hydrogen atoms in a pyridoxal 5'-phosphate enzyme.
Nat Commun, 8, 2017
|
|
6KAL
 
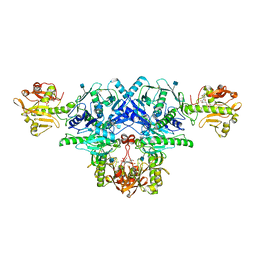 | | Crystal structure of FKRP in complex with Mg ion and CMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYTIDINE-5'-MONOPHOSPHATE, Fukutin-related protein, ... | | Authors: | Kuwabara, N. | | Deposit date: | 2019-06-23 | | Release date: | 2020-01-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of fukutin-related protein (FKRP), a ribitol-phosphate transferase related to muscular dystrophy.
Nat Commun, 11, 2020
|
|
5VKH
 
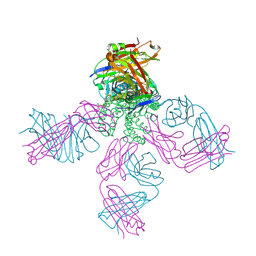 | | Closed conformation of KcsA Y82A-F103A mutant | | Descriptor: | (1S)-2-HYDROXY-1-[(NONANOYLOXY)METHYL]ETHYL MYRISTATE, Antibody Heavy Chain, Antibody Light Chain, ... | | Authors: | Cuello, L.G, Perozo, E, Cortes, D.M. | | Deposit date: | 2017-04-21 | | Release date: | 2017-12-06 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The gating cycle of a K+ channel at atomic resolution.
Elife, 6, 2017
|
|
7M5A
 
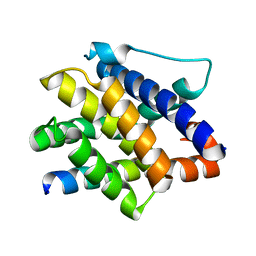 | |
3AQY
 
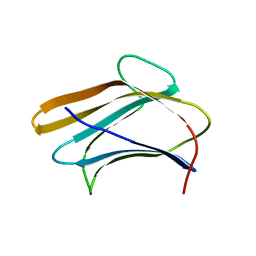 | | Crystal structure of Plodia interpunctella beta-GRP/GNBP3 N-terminal domain | | Descriptor: | Beta-1,3-glucan-binding protein | | Authors: | Kanagawa, M, Satoh, T, Ikeda, A, Adachi, Y, Ohno, N, Yamaguchi, Y. | | Deposit date: | 2010-11-22 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural insights into recognition of triple-helical beta-glucans by an insect fungal receptor
J.Biol.Chem., 286, 2011
|
|
3BLA
 
 | | Synthetic Gene Encoded DcpS bound to inhibitor DG153249 | | Descriptor: | 5-[(1S)-1-(3-chlorophenyl)ethoxy]quinazoline-2,4-diamine, Scavenger mRNA-decapping enzyme DcpS | | Authors: | Staker, B.L, Christensen, J, Stewart, L, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2007-12-10 | | Release date: | 2008-10-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | DcpS as a therapeutic target for spinal muscular atrophy.
Acs Chem.Biol., 3, 2008
|
|
5W06
 
 | | HUMAN TISSUE FACTOR IN COMPLEX WITH ANTIBODY M1587 | | Descriptor: | GLYCEROL, M1587 FAB HEAVY CHAIN, M1587 FAB LIGHT CHAIN, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T.J, Gilliland, G.L. | | Deposit date: | 2017-05-30 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into humanization of anti-tissue factor antibody 10H10.
MAbs, 10, 2018
|
|
6KLN
 
 | | F-actin of cardiac thin filament in high-calcium state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Oda, T, Yanagisawa, H, Wakabayashi, T. | | Deposit date: | 2019-07-30 | | Release date: | 2020-01-15 | | Last modified: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of cardiac thin filaments reveal the 3D architecture of troponin.
J.Struct.Biol., 209, 2020
|
|
6KN7
 
 | | Structure of human cardiac thin filament in the calcium free state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Fujii, T, Yamada, Y, Namba, K. | | Deposit date: | 2019-08-03 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Cardiac muscle thin filament structures reveal calcium regulatory mechanism.
Nat Commun, 11, 2020
|
|
3BYK
 
 | |
6KN8
 
 | | Structure of human cardiac thin filament in the calcium bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Fujii, T, Yamada, Y, Namba, K. | | Deposit date: | 2019-08-03 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cardiac muscle thin filament structures reveal calcium regulatory mechanism.
Nat Commun, 11, 2020
|
|
3BPN
 
 | | Crystal structure of the IL4-IL4R-IL13Ra ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-13 receptor alpha-1 chain, Interleukin-4, ... | | Authors: | Garcia, K.C. | | Deposit date: | 2007-12-18 | | Release date: | 2008-02-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Molecular and Structural Basis of Cytokine Receptor Pleiotropy in the Interleukin-4/13 System.
Cell(Cambridge,Mass.), 132, 2008
|
|
5VHY
 
 | | GluA2-2xGSG1L bound to ZK | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2,Germ cell-specific gene 1-like protein, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2017-04-13 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural Bases of Desensitization in AMPA Receptor-Auxiliary Subunit Complexes.
Neuron, 94, 2017
|
|
6KDK
 
 | | HIV-1 reverse transcriptase with Q151M/Y115F/F116Y:DNA:dCTP ternary complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2019-07-02 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural features in common of HBV and HIV-1 resistance against chirally-distinct nucleoside analogues entecavir and lamivudine.
Sci Rep, 10, 2020
|
|
6KAN
 
 | | Crystal structure of FKRP in complex with Ba ion | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BARIUM ION, Fukutin-related protein, ... | | Authors: | Kuwabara, N. | | Deposit date: | 2019-06-23 | | Release date: | 2020-01-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Crystal structures of fukutin-related protein (FKRP), a ribitol-phosphate transferase related to muscular dystrophy.
Nat Commun, 11, 2020
|
|
3BSH
 
 | |
5VMM
 
 | | Staphylococcus aureus IsdB bound to human hemoglobin | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, Iron-regulated cell wall-anchored protein, ... | | Authors: | Bowden, C.F, Chan, A.C, Murphy, M.E. | | Deposit date: | 2017-04-27 | | Release date: | 2017-11-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure-function analyses reveal key features in Staphylococcus aureus IsdB-associated unfolding of the heme-binding pocket of human hemoglobin.
J. Biol. Chem., 293, 2018
|
|
5VKX
 
 | |
2PNX
 
 | |
