4ZV5
 
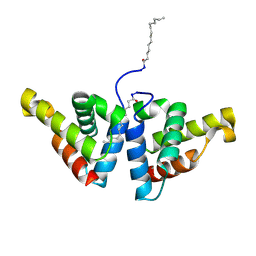 | | Crystal structure of N-myristoylated mouse mammary tumor virus matrix protein | | Descriptor: | MYRISTIC ACID, Matrix protein p10 | | Authors: | Zabransky, A, Dolezal, M, Dostal, J, Vanek, O, Hadravova, R, Stokrova, J, Brynda, J, Pichova, I. | | Deposit date: | 2015-05-18 | | Release date: | 2016-01-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Myristoylation drives dimerization of matrix protein from mouse mammary tumor virus.
Retrovirology, 13, 2016
|
|
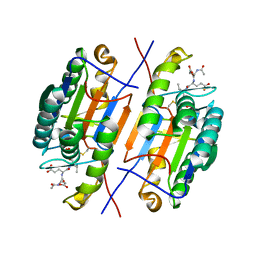
4ZVR
 
 | |
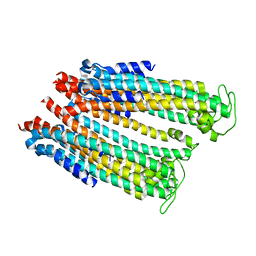
4ZWQ
 
 | |
8EGB
 
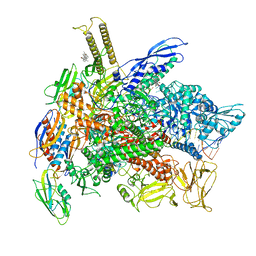 | | Cryo-EM structure of consensus elemental paused elongation complex with an unfolded TL | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Chen, J, Llewellyn, E, Landick, R, Darst, S.A. | | Deposit date: | 2022-09-12 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An ensemble of interconverting conformations of the elemental paused transcription complex creates regulatory options.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EG7
 
 | | Cryo-EM structure of pre-consensus elemental paused elongation complex | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Chen, J, Llewellyn, E, Landick, R, Darst, S.A. | | Deposit date: | 2022-09-11 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | An ensemble of interconverting conformations of the elemental paused transcription complex creates regulatory options.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EH8
 
 | | Cryo-EM structure of his-elemental paused elongation complex with a folded TL and a rotated RH-FL (1) | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Chen, J, Llewellyn, E, Landick, R, Darst, S.A. | | Deposit date: | 2022-09-13 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An ensemble of interconverting conformations of the elemental paused transcription complex creates regulatory options.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5A7D
 
 | | Tetrameric assembly of LGN with Inscuteable | | Descriptor: | INSCUTEABLE, PINS | | Authors: | Culurgioni, S, Mari, S, Bonetto, G, Gallini, S, Brennich, M, Round, A, Mapelli, M. | | Deposit date: | 2015-07-07 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Structure of the Insc:Lgn Tetramer Reveals a New Function of Lgn in Promoting Asymmetric Cell Divisions
To be Published
|
|
8EH9
 
 | | Cryo-EM structure of his-elemental paused elongation complex with a folded TL and a rotated RH-FL (2) | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Chen, J, Llewellyn, E, Landick, R, Darst, S.A. | | Deposit date: | 2022-09-13 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | An ensemble of interconverting conformations of the elemental paused transcription complex creates regulatory options.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4ZU2
 
 | | Pseudomonas aeruginosa AtuE | | Descriptor: | IODIDE ION, Putative isohexenylglutaconyl-CoA hydratase | | Authors: | Poudel, N, Pfannstiel, J, Simon, O, Walter, N, Jendrossek, D, Papageorgiou, A.C. | | Deposit date: | 2015-05-15 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Pseudomonas aeruginosa Isohexenyl Glutaconyl Coenzyme A Hydratase (AtuE) Is Upregulated in Citronellate-Grown Cells and Belongs to the Crotonase Family.
Appl.Environ.Microbiol., 81, 2015
|
|
7DPM
 
 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with MW06 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, ... | | Authors: | Wang, J, Jiao, S, Wang, R, Zhang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-12-20 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.304 Å) | | Cite: | Characterization of MW06, a human monoclonal antibody with cross-neutralization activity against both SARS-CoV-2 and SARS-CoV.
Mabs, 13, 2021
|
|
4ZVT
 
 | |
5A9E
 
 | | Cryo-electron tomography and subtomogram averaging of Rous-Sarcoma- Virus deltaMBD virus-like particles | | Descriptor: | DELTAMBD GAG PROTEIN | | Authors: | Schur, F.K.M, Dick, R.A, Hagen, W.J.H, Vogt, V.M, Briggs, J.A.G. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | The Structure of Immature-Like Rous Sarcoma Virus Gag Particles Reveals a Structural Role for the P10 Domain in Assembly.
J.Virol., 89, 2015
|
|
5ACX
 
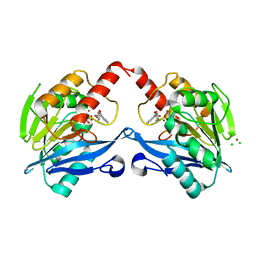 | | VIM-2-2, Discovery of novel inhibitor scaffolds against the metallo- beta-lactamase VIM-2 by SPR based fragment screening | | Descriptor: | 2-(4-fluorophenyl)carbonylbenzoic acid, BETA-LACTAMASE, CHLORIDE ION, ... | | Authors: | Christopeit, T, Carlsen, T.J.O, Helland, R, Leiros, H.K.S. | | Deposit date: | 2015-08-18 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Novel Inhibitor Scaffolds Against the Metallo-Beta-Lactamase Vim-2 by Spr Based Fragment Screening
J.Med.Chem., 58, 2015
|
|
4ZSB
 
 | |
5A3J
 
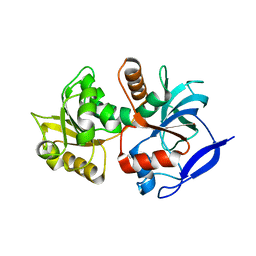 | | Crystal structure of the chloroplastic gamma-ketol reductase from Arabidopsis thaliana bound to 13-Oxo-9(Z),11(E),15(Z)- octadecatrienoic acid. | | Descriptor: | (13-oxo-9(Z),11(E),15(Z)-octadecatrienoic acid), PUTATIVE QUINONE-OXIDOREDUCTASE HOMOLOG, CHLOROPLASTIC | | Authors: | Mas-y-mas, S, Curien, G, Giustini, C, Rolland, N, Ferrer, J.L, Cobessi, D. | | Deposit date: | 2015-06-01 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.776 Å) | | Cite: | Crystal Structure of the Chloroplastic Oxoene Reductase ceQORH from Arabidopsis thaliana.
Front Plant Sci, 8, 2017
|
|
5AGV
 
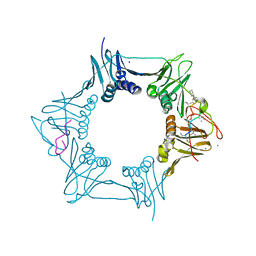 | | The sliding clamp of Mycobacterium tuberculosis in complex with a natural product. | | Descriptor: | (R,R)-2,3-BUTANEDIOL, CALCIUM ION, CYCLOHEXYL GRISELIMYCIN, ... | | Authors: | Lukat, P, Kling, A, Heinz, D.W, Mueller, R. | | Deposit date: | 2015-02-03 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Antibiotics. Targeting Dnan for Tuberculosis Therapy Using Novel Griselimycins.
Science, 348, 2015
|
|
4ZVP
 
 | |
4ZXE
 
 | | X-ray crystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. | | Descriptor: | 1,2-ETHANEDIOL, Glucanase/Chitosanase, SULFATE ION | | Authors: | Shinya, S, Oi, H, Kitaoku, Y, Ohnuma, T, Numata, T, Fukamizo, T. | | Deposit date: | 2015-05-20 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism of chitosan recognition by CBM32 carbohydrate-binding modules from a Paenibacillus sp. IK-5 chitosanase/glucanase
Biochem.J., 473, 2016
|
|
4ZSM
 
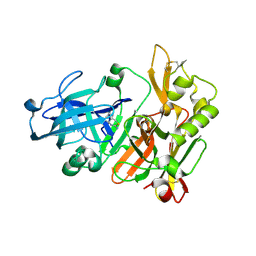 | | BACE crystal structure with bicyclic aminothiazine fragment | | Descriptor: | (4aS,8aR)-4a,5,6,7,8,8a-hexahydro-4H-3,1-benzothiazin-2-amine, Beta-secretase 1, GLYCEROL | | Authors: | Timm, D.E. | | Deposit date: | 2015-05-13 | | Release date: | 2015-06-10 | | Last modified: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Preparation and biological evaluation of conformationally constrained BACE1 inhibitors.
Bioorg.Med.Chem., 23, 2015
|
|
4ZSF
 
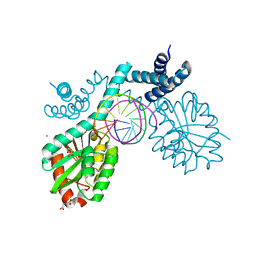 | | Crystal structure of pre-specific restriction endonuclease BsaWI-DNA complex | | Descriptor: | BsaWI endonuclease, CALCIUM ION, DNA, ... | | Authors: | Tamulaitiene, G, Rutkauskas, M, Grazulis, S, Siksnys, V. | | Deposit date: | 2015-05-13 | | Release date: | 2015-10-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Functional significance of protein assemblies predicted by the crystal structure of the restriction endonuclease BsaWI.
Nucleic Acids Res., 43, 2015
|
|
4ZT8
 
 | | Structure of the complex of type 1 ribosome inactivating protein from Momordica balsamina with a pyrimidine base, cytosine at 1.98 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-AMINOPYRIMIDIN-2(1H)-ONE, GLYCEROL, ... | | Authors: | Yamini, S, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-05-14 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein fromMomordica balsaminawith cytosine, cytidine, and cytidine diphosphate.
Biochem Biophys Rep, 4, 2015
|
|
4ZVS
 
 | |
8ET1
 
 | |
4ZRZ
 
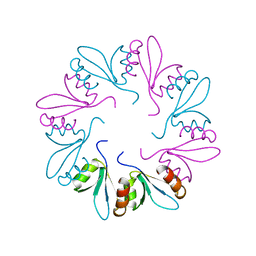 | | PlyCB mutant R66E | | Descriptor: | PlyCB | | Authors: | Gallagher, D.T, Nelson, D.C, Shen, Y. | | Deposit date: | 2015-05-12 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A bacteriophage endolysin that eliminates intracellular streptococci.
Elife, 5, 2016
|
|
4ZS8
 
 | | Crystal structure of ligand-free, full length DasR | | Descriptor: | 1,2-ETHANEDIOL, HTH-type transcriptional repressor DasR | | Authors: | Fillenberg, S.B, Muller, Y.A. | | Deposit date: | 2015-05-13 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of the Global Regulator DasR from Streptomyces coelicolor: Implications for the Allosteric Regulation of GntR/HutC Repressors.
Plos One, 11, 2016
|
|
