5NS8
 
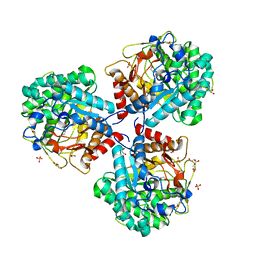 | | Crystal structure of beta-glucosidase BglM-G1 mutant H75R from marine metagenome in complex with inhibitor 1-Deoxynojirimycin | | Descriptor: | 1-DEOXYNOJIRIMYCIN, GLYCEROL, SULFATE ION, ... | | Authors: | Mhaindarkar, D.C, Gasper, R, Lupilova, N, Leichert, L.I, Hofmann, E. | | Deposit date: | 2017-04-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Loss of a conserved salt bridge in bacterial glycosyl hydrolase BgIM-G1 improves substrate binding in temperate environments.
Commun Biol, 1, 2018
|
|
7DN7
 
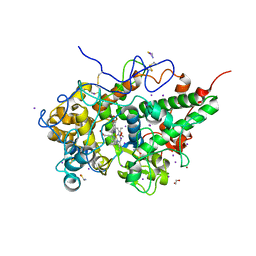 | | Crystal structure of ternary complexes of lactoperoxidase with hydrogen peroxide at 1.70 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, P.K, Singh, A.K, Singh, R.P, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2020-12-09 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a ternary complex of lactoperoxidase with iodide and hydrogen peroxide at 1.77 angstrom resolution.
J.Inorg.Biochem., 220, 2021
|
|
7DLQ
 
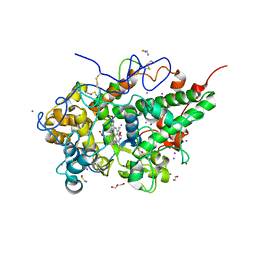 | | CRYSTAL STRUCTURE OF THE COMPLEX OF LACTOPEROXIDASE WITH HYDROGEN PEROXIDE AT 1.77A RESOLUTION | | Descriptor: | 1,2-ETHANEDIOL, 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, P.K, Sharma, P, Bhushan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2020-11-29 | | Release date: | 2020-12-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | Structure of a ternary complex of lactoperoxidase with iodide and hydrogen peroxide at 1.77 angstrom resolution.
J.Inorg.Biochem., 220, 2021
|
|
7DN6
 
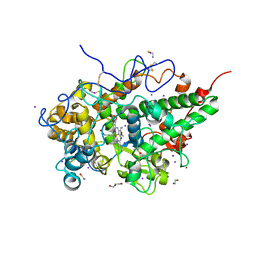 | | Crystal structure of bovine lactoperoxidase with hydrogen peroxide trapped between heme iron and his109 at 1.69 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, P.K, Singh, A.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2020-12-08 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Structure of a ternary complex of lactoperoxidase with iodide and hydrogen peroxide at 1.77 angstrom resolution.
J.Inorg.Biochem., 220, 2021
|
|
5FSD
 
 | | 1.75 A resolution 2,5-dihydroxybenzensulfonate inhibited Sporosarcina pasteurii urease | | Descriptor: | 1,2-ETHANEDIOL, 2,5-dihydroxybenzenesulfonic acid, HYDROXIDE ION, ... | | Authors: | Mazzei, L, Cianci, M, Musiani, F, Ciurli, S. | | Deposit date: | 2016-01-04 | | Release date: | 2016-03-23 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Inactivation of Urease by 1,4-Benzoquinone: Chemistry at the Protein Surface.
Dalton Trans, 45, 2016
|
|
3LWZ
 
 | | 1.65 Angstrom Resolution Crystal Structure of Type II 3-Dehydroquinate Dehydratase (aroQ) from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, 3-dehydroquinate dehydratase, BETA-MERCAPTOETHANOL, ... | | Authors: | Minasov, G, Light, S.H, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-02-24 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | 1.65 Angstrom Resolution Crystal Structure of Type II 3-Dehydroquinate Dehydratase (aroQ) from Yersinia pestis.
TO BE PUBLISHED
|
|
5I2A
 
 | | 1,2-propanediol Dehydration in Roseburia inulinivorans; Structural Basis for Substrate and Enantiomer Selectivity | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Diol-dehydratase | | Authors: | LaMattina, J.W, Reitzer, P, Kapoor, S, Galzerani, F, Koch, D.J, Gouvea, I.E, Lanzilotta, W.N. | | Deposit date: | 2016-02-08 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 1,2-Propanediol Dehydration in Roseburia inulinivorans: STRUCTURAL BASIS FOR SUBSTRATE AND ENANTIOMER SELECTIVITY.
J.Biol.Chem., 291, 2016
|
|
9IT8
 
 | | Crystal structure of the ternary complex of lactoperoxidase with nitric oxide and nitrite ion at 1.95 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Maurya, A, Ahmad, N, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2024-07-19 | | Release date: | 2024-09-11 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Structure of the Complex of Lactoperoxidase With Nitric Oxide at 1.95 angstrom Resolution.
Proteins, 93, 2025
|
|
6ELI
 
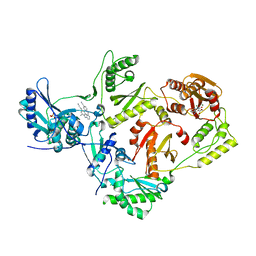 | | Structure of HIV-1 reverse transcriptase (RT) in complex with rilpivirine and an RNase H inhibitor XZ462 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, Gag-Pol polyprotein, ... | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2017-09-29 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Developing and Evaluating Inhibitors against the RNase H Active Site of HIV-1 Reverse Transcriptase.
J. Virol., 92, 2018
|
|
7C7K
 
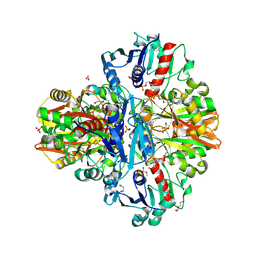 | | Crystal Structure of Thioacyl-Glyceraldehyde-3-phosphate dehydrogenase 1(GAPDH 1) from Escherichia coli at 1.77 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, ACETONE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-26 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
6FSE
 
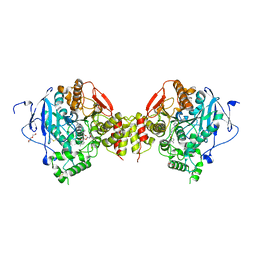 | | Mus musculus acetylcholinesterase in complex with 1-(4-(4-Ethylpiperazin-1-yl)piperidin-1-yl)-2-((4'-methoxy-[1,1'-biphenyl]-4-yl)oxy)ethanone dihydrochloride (15) | | Descriptor: | 1-[4-(4-ethylpiperazin-1-yl)piperidin-1-yl]-2-[4-(4-methoxyphenyl)phenoxy]ethanone, 2-(2-METHOXYETHOXY)ETHANOL, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ... | | Authors: | Knutsson, S, Engdahl, C, Kumari, R, Kindahl, T, Forsgren, N, Lindgren, C, Kitur, S, Kamau, L, Ekstrom, F, Linusson, A. | | Deposit date: | 2018-02-19 | | Release date: | 2018-10-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Noncovalent Inhibitors of Mosquito Acetylcholinesterase 1 with Resistance-Breaking Potency.
J.Med.Chem., 61, 2018
|
|
6DTX
 
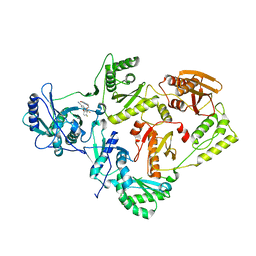 | | Wildtype HIV-1 Reverse Transcriptase in complex with JLJ 578 | | Descriptor: | 8-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-fluorophenoxy}-6-fluoroindolizine-2-carbonitrile, GLYCEROL, Reverse transcriptase/ribonuclease H, ... | | Authors: | Sasaki, T, Gannam, Z.T.K, Anderson, K.S, Jorgensen, W.L, Lee, W. | | Deposit date: | 2018-06-18 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.327 Å) | | Cite: | Molecular and cellular studies evaluating a potent 2-cyanoindolizine catechol diether NNRTI targeting wildtype and Y181C mutant HIV-1 reverse transcriptase.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6DTW
 
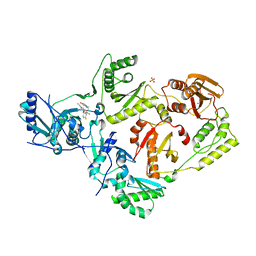 | | HIV-1 Reverse Transcriptase Y181C Mutant in complex with JLJ 578 | | Descriptor: | 8-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-fluorophenoxy}-6-fluoroindolizine-2-carbonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Sasaki, T, Gannam, Z.T.K, Anderson, K.S, Jorgensen, W.L, Lee, W. | | Deposit date: | 2018-06-18 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.742 Å) | | Cite: | Molecular and cellular studies evaluating a potent 2-cyanoindolizine catechol diether NNRTI targeting wildtype and Y181C mutant HIV-1 reverse transcriptase.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6C0O
 
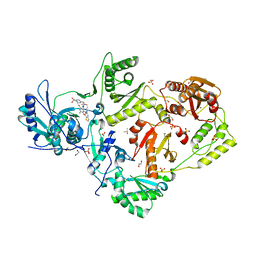 | | Crystal structure of HIV-1 K103N mutant reverse transcriptase in complex with non-nucleoside inhibitor 25a | | Descriptor: | 1,2-ETHANEDIOL, 4-({4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}thieno[3,2-d]pyrimidin-2-yl)amino]piperidin-1-yl}methyl)benzene-1-sulfonamide, DIMETHYL SULFOXIDE, ... | | Authors: | Yang, Y, Nguyen, L.A, Smithline, Z.B, Steitz, T.A. | | Deposit date: | 2018-01-01 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors.
Elife, 7, 2018
|
|
7C5G
 
 | | Crystal Structure of C150S mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli complexed with PO4 at 1.98 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Zhang, L, Liu, M.R, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5L
 
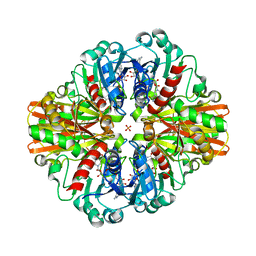 | | Crystal Structure of C150S mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli at 2.1 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5J
 
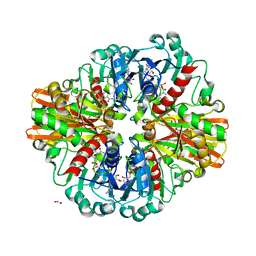 | | Crystal Structure of C150A mutant of Glyceraldehyde-3-phosphate dehydrogenase1 from Escherichia coli at 1.98 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5M
 
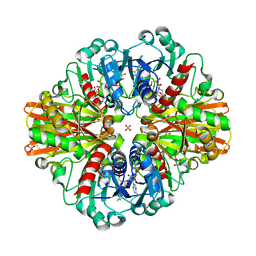 | | Crystal Structure of C150A+H177A mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli complexed with G3P at 1.8 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 3-PHOSPHOGLYCERIC ACID, GLYCERALDEHYDE-3-PHOSPHATE, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
6C0N
 
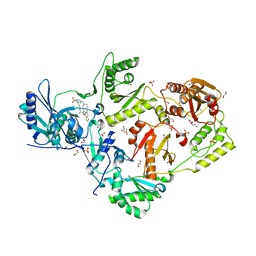 | | Crystal structure of HIV-1 reverse transcriptase in complex with non-nucleoside inhibitor 25a | | Descriptor: | 1,2-ETHANEDIOL, 4-({4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}thieno[3,2-d]pyrimidin-2-yl)amino]piperidin-1-yl}methyl)benzene-1-sulfonamide, DIMETHYL SULFOXIDE, ... | | Authors: | Yang, Y, Nguyen, L.A, Smithline, Z.B, Steitz, T.A. | | Deposit date: | 2018-01-01 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors.
Elife, 7, 2018
|
|
6C0P
 
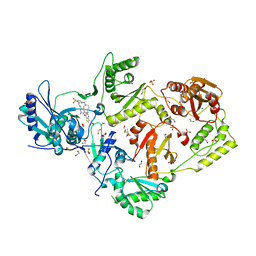 | | Crystal structure of HIV-1 E138K mutant reverse transcriptase in complex with non-nucleoside inhibitor 25a | | Descriptor: | 1,2-ETHANEDIOL, 4-({4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}thieno[3,2-d]pyrimidin-2-yl)amino]piperidin-1-yl}methyl)benzene-1-sulfonamide, DIMETHYL SULFOXIDE, ... | | Authors: | Yang, Y, Nguyen, L.A, Smithline, Z.B, Steitz, T.A. | | Deposit date: | 2018-01-01 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors.
Elife, 7, 2018
|
|
6C0L
 
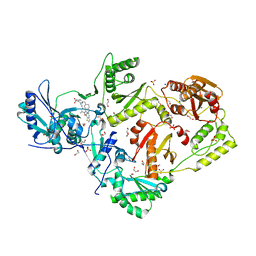 | | Crystal structure of HIV-1 E138K mutant reverse transcriptase in complex with non-nucleoside inhibitor K-5a2 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{[4-(4-cyano-2,6-dimethylphenoxy)thieno[3,2-d]pyrimidin-2-yl]amino}piperidin-1-yl)methyl]benzene-1-sulfonamide, MAGNESIUM ION, ... | | Authors: | Yang, Y, Nguyen, L.A, Smithline, Z.B, Steitz, T.A. | | Deposit date: | 2018-01-01 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors.
Elife, 7, 2018
|
|
6C0J
 
 | | Crystal structure of HIV-1 reverse transcriptase in complex with non-nucleoside inhibitor K-5a2 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{[4-(4-cyano-2,6-dimethylphenoxy)thieno[3,2-d]pyrimidin-2-yl]amino}piperidin-1-yl)methyl]benzene-1-sulfonamide, MAGNESIUM ION, ... | | Authors: | Yang, Y, Nguyen, L.A, Smithline, Z.B, Steitz, T.A. | | Deposit date: | 2018-01-01 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors.
Elife, 7, 2018
|
|
6C0K
 
 | | Crystal structure of HIV-1 K103N mutant reverse transcriptase in complex with non-nucleoside inhibitor K-5a2 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{[4-(4-cyano-2,6-dimethylphenoxy)thieno[3,2-d]pyrimidin-2-yl]amino}piperidin-1-yl)methyl]benzene-1-sulfonamide, DIMETHYL SULFOXIDE, ... | | Authors: | Yang, Y, Nguyen, L.A, Smithline, Z.B, Steitz, T.A. | | Deposit date: | 2018-01-01 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.958 Å) | | Cite: | Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors.
Elife, 7, 2018
|
|
6C0R
 
 | | Crystal structure of HIV-1 K103N/Y181C mutant reverse transcriptase in complex with non-nucleoside inhibitor 25a | | Descriptor: | 1,2-ETHANEDIOL, 4-({4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}thieno[3,2-d]pyrimidin-2-yl)amino]piperidin-1-yl}methyl)benzene-1-sulfonamide, DIMETHYL SULFOXIDE, ... | | Authors: | Yang, Y, Nguyen, L.A, Smithline, Z.B, Steitz, T.A. | | Deposit date: | 2018-01-02 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors.
Elife, 7, 2018
|
|
6DNQ
 
 | | HBZ77 in complex with KIX and c-Myb | | Descriptor: | 1,2-ETHANEDIOL, BZIP factor, CREB-binding protein, ... | | Authors: | Yang, K, Wright, P.E, Stanfield, R.L. | | Deposit date: | 2018-06-07 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for cooperative regulation of KIX-mediated transcription pathways by the HTLV-1 HBZ activation domain.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
