8SG9
 
 | | CCT G beta 5 complex closed state 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-12 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SGC
 
 | | CCT G beta 5 complex closed state 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-12 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
4PG7
 
 | |
8SGQ
 
 | | CCT G beta 5 complex intermediate state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-12 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SHN
 
 | | CCT-G beta 5 complex closed state 6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SG8
 
 | | CCT G beta 5 complex closed state 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-11 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SHG
 
 | | CCT G beta 5 complex closed state 9 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SHF
 
 | | CCT-G beta 5 complex closed state 7 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-14 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SHQ
 
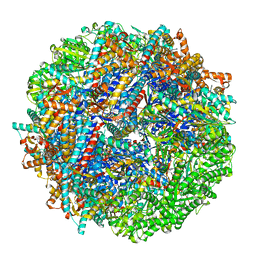 | | CCT G beta 5 complex closed state 12 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
2TRX
 
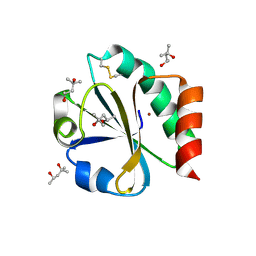 | | CRYSTAL STRUCTURE OF THIOREDOXIN FROM ESCHERICHIA COLI AT 1.68 ANGSTROMS RESOLUTION | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, THIOREDOXIN | | Authors: | Katti, S.K, Lemaster, D.M, Eklund, H. | | Deposit date: | 1990-03-19 | | Release date: | 1991-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of thioredoxin from Escherichia coli at 1.68 A resolution.
J.Mol.Biol., 212, 1990
|
|
6EQL
 
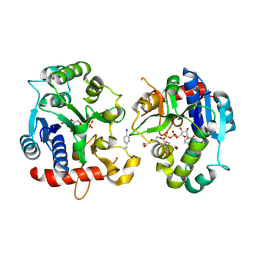 | | Crystal Structure of Human Glycogenin-1 (GYG1) Tyr195pIPhe mutant complexed with manganese and UDP | | Descriptor: | 1,2-ETHANEDIOL, Glycogenin-1, MANGANESE (II) ION, ... | | Authors: | Bailey, H.J, Kopec, J, Bilyard, M.K, Bezerra, G.A, Seo Lee, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Davis, B.G, Yue, W.W. | | Deposit date: | 2017-10-13 | | Release date: | 2017-12-20 | | Last modified: | 2018-11-14 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Palladium-mediated enzyme activation suggests multiphase initiation of glycogenesis.
Nature, 563, 2018
|
|
8C9Z
 
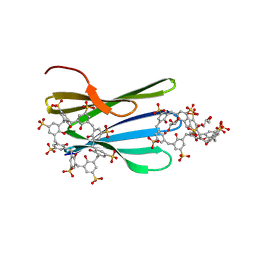 | | The RSL - sulfonato-calix[8]arene complex, H32 form, citrate pH 6.0 | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, beta-D-fructopyranose, ... | | Authors: | Mockler, N.M, Ramberg, K, Crowley, P.B. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Protein-macrocycle polymorphism: crystal form IV of the Ralstonia solanacearum lectin-sulfonato-calix[8]arene complex.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8C9Y
 
 | | The MK-RSL - sulfonato-calix[8]arene complex, H32 form | | Descriptor: | GLYCEROL, Putative fucose-binding lectin protein, beta-D-fructopyranose, ... | | Authors: | Mockler, N.M, Ramberg, K, Crowley, P.B. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Protein-macrocycle polymorphism: crystal form IV of the Ralstonia solanacearum lectin-sulfonato-calix[8]arene complex.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
6RST
 
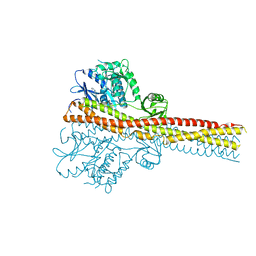 | | TBK1 in complex with inhibitor compound 24 | | Descriptor: | 1-[4-[(1~{R})-1-[2-[[5-[1-(cyclopropylmethyl)pyrazol-4-yl]-1~{H}-benzimidazol-2-yl]amino]pyridin-4-yl]ethyl]piperazin-1-yl]-3,3,3-tris(fluoranyl)propan-1-one, Serine/threonine-protein kinase TBK1 | | Authors: | Panne, D, Hillig, R.C, Rengachari, S. | | Deposit date: | 2019-05-22 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Discovery of BAY-985, a Highly Selective TBK1/IKK epsilon Inhibitor.
J.Med.Chem., 63, 2020
|
|
6RT9
 
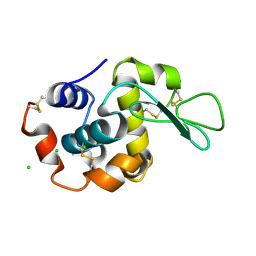 | |
6UJV
 
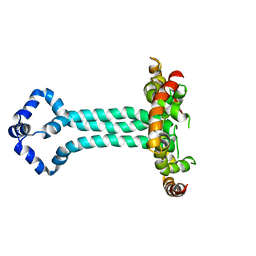 | | Model of the HIV-1 gp41 membrane-proximal external region, transmembrane domain and cytoplasmic tail (LLP2) | | Descriptor: | Envelope glycoprotein GP41 | | Authors: | Piai, A, Fu, Q, Cai, Y, Ghantous, F, Xiao, T, Shaik, M.M, Peng, H, Rits-Volloch, S, Liu, Z, Chen, W, Seaman, M.S, Chen, B, Chou, J.J. | | Deposit date: | 2019-10-03 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of transmembrane coupling of the HIV-1 envelope glycoprotein.
Nat Commun, 11, 2020
|
|
8CJ2
 
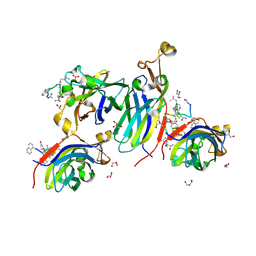 | | Urea-based foldamer inhibitor c3u_5 chimera in complex with ASF1 histone chaperone | | Descriptor: | GLYCEROL, Histone chaperone ASF1A, SULFATE ION, ... | | Authors: | Perrin, M.E, Li, B, Mbianda, J, Ropars, V, Legrand, P, Douat, C, Ochsenbein, F, Guichard, G. | | Deposit date: | 2023-02-11 | | Release date: | 2023-07-05 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.127 Å) | | Cite: | Unexpected binding modes of inhibitors to the histone chaperone ASF1 revealed by a foldamer scanning approach.
Chem.Commun.(Camb.), 59, 2023
|
|
8CJ1
 
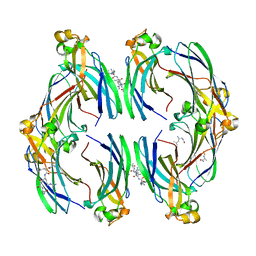 | | Urea-based foldamer inhibitor c3u_3 chimera in complex with ASF1 histone chaperone | | Descriptor: | Histone chaperone ASF1A, c3u_3 chimera inhibitor of histone chaperone ASF1 | | Authors: | Perrin, M.E, Li, B, Mbianda, J, Ropars, V, Legrand, P, Douat, C, Ochsenbein, F, Guichard, G. | | Deposit date: | 2023-02-11 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.564 Å) | | Cite: | Unexpected binding modes of inhibitors to the histone chaperone ASF1 revealed by a foldamer scanning approach.
Chem.Commun.(Camb.), 59, 2023
|
|
6RWH
 
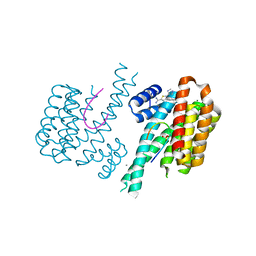 | | Fragment AZ-007 binding at a primary and secondary binding site of the the p53pT387/14-3-3 sigma complex | | Descriptor: | 14-3-3 protein sigma, 5-(1~{H}-imidazol-5-yl)-4-phenyl-thiophene-2-carboximidamide, CALCIUM ION, ... | | Authors: | Leysen, S, Guillory, X, Wolter, M, Genet, S, Somsen, B, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-06-05 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
7ZGU
 
 | | Human NLRP3-deltaPYD hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, NACHT, LRR and PYD domains-containing protein 3 | | Authors: | Raisch, T, Machtens, D.A, Bresch, I.B, Eberhage, J, Prumbaum, D, Reubold, T.F, Raunser, S, Eschenburg, S. | | Deposit date: | 2022-04-04 | | Release date: | 2022-05-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the NEK7-independent NLRP3 inflammasome
To Be Published
|
|
8HT1
 
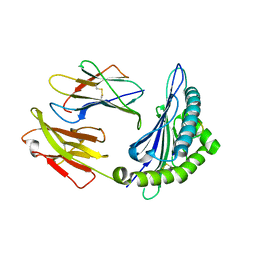 | |
8SPC
 
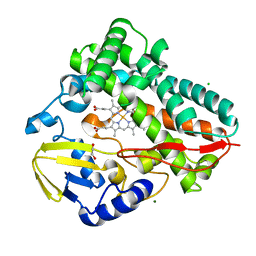 | |
7JT6
 
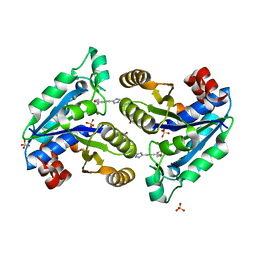 | | Mycobacterium tuberculosis dethiobiotin synthetase in complex with Tetrazole 2 | | Descriptor: | ATP-dependent dethiobiotin synthetase BioD, GLYCEROL, SULFATE ION, ... | | Authors: | Pederick, J.P, Bean, J.H, Bruning, J.B. | | Deposit date: | 2020-08-17 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of Mycobacterium tuberculosis Dethiobiotin Synthase ( Mt DTBS): Toward Next-Generation Antituberculosis Agents.
Acs Chem.Biol., 16, 2021
|
|
8HSO
 
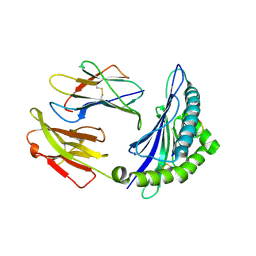 | |
1OPQ
 
 | | NMR structure of unmethylated GATC site | | Descriptor: | 5'-D(*CP*GP*CP*AP*GP*AP*TP*CP*TP*CP*GP*C)-3', 5'-D(*GP*CP*GP*AP*GP*AP*TP*CP*TP*GP*CP*G)-3' | | Authors: | Bae, S.-H, Cheong, H.-K, Kang, S, Hwang, D.S, Cheong, C, Choi, B.-S. | | Deposit date: | 2003-03-06 | | Release date: | 2004-04-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of hemimethylated GATC sites: implications for DNA-SeqA recognition
J.Biol.Chem., 278, 2003
|
|
