2LTU
 
 | |
1Z8D
 
 | | Crystal Structure of Human Muscle Glycogen Phosphorylase a with AMP and Glucose | | Descriptor: | ADENINE, ADENOSINE MONOPHOSPHATE, Glycogen phosphorylase, ... | | Authors: | Lukacs, C.M, Oikonomakos, N.G, Crowther, R.L, Hong, L.N, Kammlott, R.U, Levin, W, Li, S, Liu, C.M, Lucas-McGady, D, Pietranico, S, Reik, L. | | Deposit date: | 2005-03-30 | | Release date: | 2006-03-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of human muscle glycogen phosphorylase a with bound glucose and AMP: An intermediate conformation with T-state and R-state features.
Proteins, 63, 2006
|
|
1FSA
 
 | | THE T-STATE STRUCTURE OF LYS 42 TO ALA MUTANT OF THE PIG KIDNEY FRUCTOSE 1,6-BISPHOSPHATASE EXPRESSED IN E. COLI | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, FRUCTOSE 1,6-BISPHOSPHATASE, ... | | Authors: | Lu, G, Stec, B, Giroux, E, Kantrowitz, E.R. | | Deposit date: | 1996-08-24 | | Release date: | 1997-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evidence for an active T-state pig kidney fructose 1,6-bisphosphatase: interface residue Lys-42 is important for allosteric inhibition and AMP cooperativity.
Protein Sci., 5, 1996
|
|
4EEI
 
 | | Crystal Structure of Adenylosuccinate Lyase from Francisella tularensis Complexed with AMP and Succinate | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Adenylosuccinate lyase, ... | | Authors: | Maltseva, N, Kim, Y, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-28 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.921 Å) | | Cite: | Crystal Structure of Adenylosuccinate Lyase from Francisella tularensis Complexed with AMP and Succinate
To be Published
|
|
2OUR
 
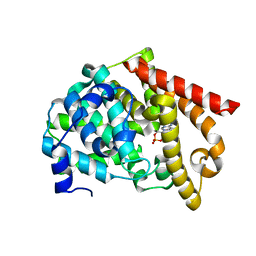 | | crystal structure of PDE10A2 mutant D674A in complex with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, MAGNESIUM ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Wang, H.C, Liu, Y.D, Hou, J, Zheng, M.Y, Robinson, H. | | Deposit date: | 2007-02-12 | | Release date: | 2007-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | From the Cover: Structural insight into substrate specificity of phosphodiesterase 10.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3AW8
 
 | | Crystal structure of N5-carboxyaminoimidazole ribonucleotide synthetase from Thermus thermophilus HB8 | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, Phosphoribosylaminoimidazole carboxylase, ... | | Authors: | Okada, K, Tsunoda, S, Taka, H, Baba, S, Kanagawa, M, Nakagawa, N, Ebihara, A, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-03-15 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of N5-carboxyaminoimidazole ribonucleotide synthetase, PurK, from thermophilic bacteria
To be Published
|
|
2PW3
 
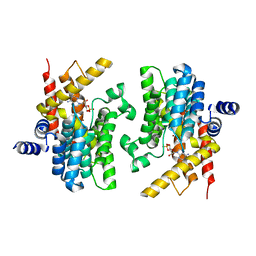 | | Structure of the PDE4D-cAMP complex | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ZINC ION, cAMP-specific 3',5'-cyclic phosphodiesterase 4D | | Authors: | Wang, H, Robinson, H, Ke, H. | | Deposit date: | 2007-05-10 | | Release date: | 2007-10-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The molecular basis for different recognition of substrates by phosphodiesterase families 4 and 10.
J.Mol.Biol., 371, 2007
|
|
6BLJ
 
 | |
5F9Y
 
 | |
4EUM
 
 | | Crystal structure of a sugar kinase (Target EFI-502132) from Oceanicola granulosus with bound AMP, crystal form II | | Descriptor: | 2-dehydro-3-deoxygluconokinase, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a sugar kinase (Target EFI-502132) from Oceanicola granulosus with bound AMP, crystal form II
To be Published
|
|
7AGY
 
 | |
7AHT
 
 | |
7AGW
 
 | |
7FTL
 
 | | Crystal Structure of human cyclic GMP-AMP synthase in complex with 8-chloro-2-(2-hydroxyphenyl)quinoline-4-carboxylic acid | | Descriptor: | (2P)-8-chloro-2-(2-hydroxyphenyl)quinoline-4-carboxylic acid, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Leibrock, L, Benz, J, Groebke-Zbinden, K, Brunner, M, Rudolph, M.G. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a human cyclic GMP-AMP synthase complex
To be published
|
|
7FTU
 
 | | Crystal Structure of human cyclic GMP-AMP synthase in complex with (Z)-N-[4-(4-chlorophenyl)sulfonylphenyl]-2-cyano-3-hydroxy-3-(5-methyl-1,2-oxazol-4-yl)prop-2-enamide | | Descriptor: | (2Z)-N-[4-(4-chlorobenzene-1-sulfonyl)phenyl]-2-cyano-3-hydroxy-3-(5-methyl-1,2-oxazol-4-yl)prop-2-enamide, ADENOSINE-5'-TRIPHOSPHATE, Cyclic GMP-AMP synthase, ... | | Authors: | Leibrock, L, Benz, J, Groebke-Zbinden, K, Rudolph, M.G. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of a human cyclic GMP-AMP synthase complex
To be published
|
|
7FTX
 
 | |
7FU5
 
 | | Crystal Structure of human cyclic GMP-AMP synthase | | Descriptor: | (8S)-2-{[(3-fluoro[1,1'-biphenyl]-4-yl)methyl]amino}-5-propyl[1,2,4]triazolo[1,5-a]pyrimidin-7(4H)-one, Cyclic GMP-AMP synthase, SULFATE ION, ... | | Authors: | Leibrock, L, Benz, J, Groebke-Zbinden, K, Rudolph, M.G. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal Structure of a human cyclic GMP-AMP synthase complex
To be published
|
|
7FU7
 
 | |
2CGP
 
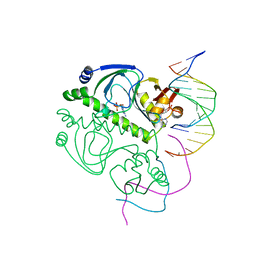 | | CATABOLITE GENE ACTIVATOR PROTEIN/DNA COMPLEX, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, DNA (5'-D(*AP*TP*TP*AP*AP*TP*GP*TP*GP*AP*CP*AP*TP*AP*T)-3'), DNA (5'-D(*GP*TP*CP*AP*CP*AP*TP*TP*AP*AP*T)-3'), ... | | Authors: | Passner, J.M, Steitz, T.A. | | Deposit date: | 1997-01-31 | | Release date: | 1998-02-04 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of a CAP-DNA complex having two cAMP molecules bound to each monomer.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
5B8H
 
 | |
6FTF
 
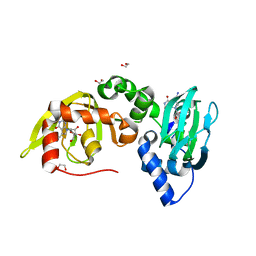 | | Regulatory subunit of a cAMP-independent protein kinase A from Trypanosoma cruzi at 1.09 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 7-cyano-7-deazainosine, Protein kinase A regulatory subunit, ... | | Authors: | Volpato Santos, Y, Lorentzen, E, Basquin, J, Boshart, M. | | Deposit date: | 2018-02-21 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.09151936 Å) | | Cite: | Nucleoside analogue activators of cyclic AMP-independent protein kinase A of Trypanosoma.
Nat Commun, 10, 2019
|
|
7D4U
 
 | | ATP complex with double mutant cyclic trinucleotide synthase CdnD | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cyclic AMP-AMP-GMP synthase | | Authors: | Yang, C.-S, Hou, M.-H, Tsai, C.-L, Wang, Y.-C, Ko, T.-P, Chen, Y. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and functional implication of a bacterial cyclic AMP-AMP-GMP synthetase.
Nucleic Acids Res., 49, 2021
|
|
7D4S
 
 | | apo-form cyclic trinucleotide synthase CdnD | | Descriptor: | Cyclic AMP-AMP-GMP synthase, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION | | Authors: | Yang, C.-S, Hou, M.-H, Tsai, C.-L, Wang, Y.-C, Ko, T.-P, Chen, Y. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure and functional implication of a bacterial cyclic AMP-AMP-GMP synthetase.
Nucleic Acids Res., 49, 2021
|
|
7FTI
 
 | | Crystal Structure of human cyclic GMP-AMP synthase in complex with 5-(4-fluorophenyl)-2-methylpyrazole-3-carboxylic acid | | Descriptor: | 3-(4-fluorophenyl)-1-methyl-1H-pyrazole-5-carboxylic acid, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Leibrock, L, Benz, J, Groebke-Zbinden, K, Brunner, M, Rudolph, M.G. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a human cyclic GMP-AMP synthase complex
To be published
|
|
7FTJ
 
 | |
