3KW4
 
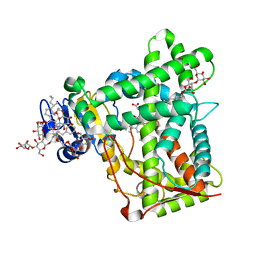 | | Crystal structure of cytochrome 2B4 in complex with the anti-platelet drug ticlopidine | | Descriptor: | 2-{[(3alpha,5alpha,7alpha,8alpha,10alpha,12alpha,17alpha)-3,12-bis{2-[(4-O-alpha-D-glucopyranosyl-beta-D-glucopyranosyl)oxy]ethoxy}cholan-7-yl]oxy}ethyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B4, ... | | Authors: | Gay, S.C, Maekawa, K, Roberts, A.G, Hong, W.-X, Zhang, Q, Stout, C.D, Halpert, J.R. | | Deposit date: | 2009-11-30 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structures of cytochrome P450 2B4 complexed with the antiplatelet drugs ticlopidine and clopidogrel.
Biochemistry, 49, 2010
|
|
3LNP
 
 | | Crystal Structure of Amidohydrolase family Protein OLEI01672_1_465 from Oleispira antarctica | | Descriptor: | ACETIC ACID, Amidohydrolase family Protein OLEI01672_1_465, CALCIUM ION, ... | | Authors: | Kim, Y, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-02 | | Release date: | 2010-02-16 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
2OHG
 
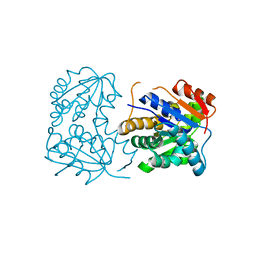 | |
2HPM
 
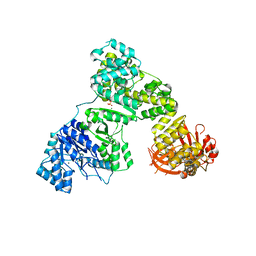 | | Eubacterial and Eukaryotic Replicative DNA Polymerases are not Homologous: X-ray Structure of DNA Polymerase III | | Descriptor: | CHLORIDE ION, DNA Polymerase III alpha subunit, MAGNESIUM ION, ... | | Authors: | Bailey, S, Wing, R.A, Steitz, T.A. | | Deposit date: | 2006-07-17 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | The Structure of T. aquaticus DNA Polymerase III Is Distinct from Eukaryotic Replicative DNA Polymerases.
Cell(Cambridge,Mass.), 126, 2006
|
|
2W3T
 
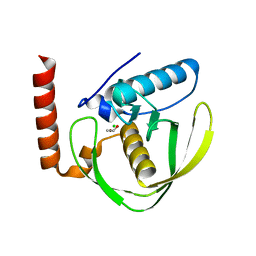 | | Chloro complex of the Ni-Form of E.coli deformylase | | Descriptor: | CHLORIDE ION, ETHANOL, NICKEL (II) ION, ... | | Authors: | Ngo, Y.H.T, Palm, G.J, Hinrichs, W. | | Deposit date: | 2008-11-14 | | Release date: | 2009-12-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure of the Ni(II) Complex of Escherichia Coli Peptide Deformylase and Suggestions on Deformylase Activities Depending on Different Metal(II) Centres.
J.Biol.Inorg.Chem., 15, 2010
|
|
2HSN
 
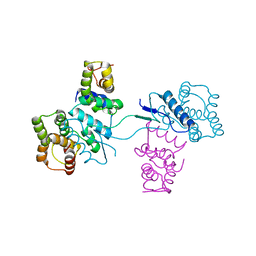 | | Structural basis of yeast aminoacyl-tRNA synthetase complex formation revealed by crystal structures of two binary sub-complexes | | Descriptor: | GU4 nucleic-binding protein 1, Methionyl-tRNA synthetase, cytoplasmic | | Authors: | Simader, H, Koehler, C, Basquin, J, Suck, D. | | Deposit date: | 2006-07-22 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of yeast aminoacyl-tRNA synthetase complex formation revealed by crystal structures of two binary sub-complexes.
Nucleic Acids Res., 34, 2006
|
|
2HPV
 
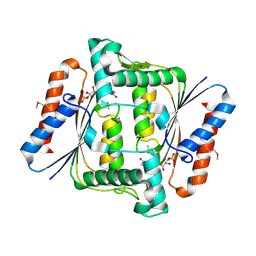 | | Crystal structure of FMN-Dependent azoreductase from Enterococcus faecalis | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase | | Authors: | Liu, Z.J, Chen, L, Chen, H, Rose, J, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-07-17 | | Release date: | 2006-09-12 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Fmn-Dependent Azoreductase from Enterococcus faecalis at 2.00 A resolution
To be Published
|
|
2HSM
 
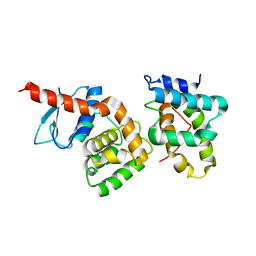 | |
2HRT
 
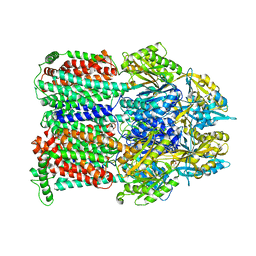 | | Asymmetric structure of trimeric AcrB from Escherichia coli | | Descriptor: | Acriflavine resistance protein B, CITRATE ANION | | Authors: | Seeger, M.A, Schiefner, A, Eicher, T, Verrey, F, Diederichs, K, Pos, K.M. | | Deposit date: | 2006-07-20 | | Release date: | 2006-09-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Asymmetry of AcrB Trimer Suggests a Peristaltic Pump Mechanism.
Science, 313, 2006
|
|
2QT3
 
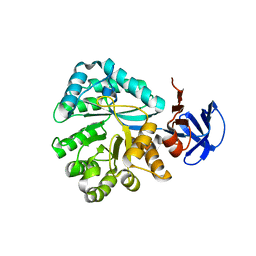 | | Crystal structure of N-Isopropylammelide isopropylaminohydrolase AtzC from Pseudomonas sp. strain ADP complexed with Zn | | Descriptor: | N-isopropylammelide isopropyl amidohydrolase, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Seffernick, J, Wackett, L.P, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-01 | | Release date: | 2007-09-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal structure of N-Isopropylammelide isopropylaminohydrolase AtzC from Pseudomonas sp. strain ADP complexed with Zn.
To be Published
|
|
2QJ8
 
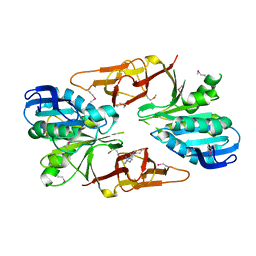 | |
2L5N
 
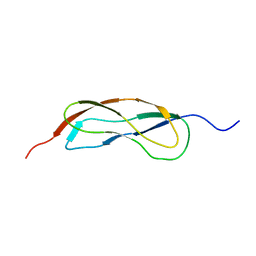 | | NMR Structure of YbbR family protein Dhaf_0833 (residues 32-118) from Desulfitobacterium hafniense DCB-2: Northeast Structural Genomics Consortium target DhR29B | | Descriptor: | YbbR family protein | | Authors: | Cort, J.R, Barb, A.W, Lee, H, Ramelot, T.A, Yang, Y, Belote, R.L, Ciccosanti, C.R, Haleema, J, Acton, T.B, Xiao, R.R, Everett, J.K, Montelione, G.T, Prestegard, J.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-11-02 | | Release date: | 2010-12-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures of domains I and IV from YbbR are representative of a widely distributed protein family.
Protein Sci., 20, 2011
|
|
2P8B
 
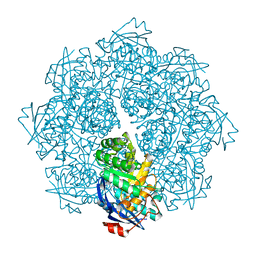 | | Crystal structure of N-succinyl Arg/Lys racemase from Bacillus cereus ATCC 14579 complexed with N-succinyl Lys. | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme family protein, N-SUCCINYL LYSINE | | Authors: | Fedorov, A.A, Song, L, Fedorov, E.V, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2007-03-22 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Prediction and assignment of function for a divergent N-succinyl amino acid racemase.
Nat.Chem.Biol., 3, 2007
|
|
2JJP
 
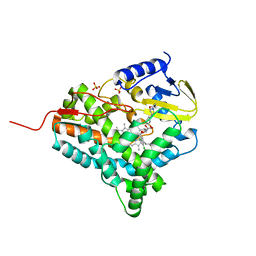 | | Structure of cytochrome P450 EryK in complex with inhibitor ketoconazole (KC) | | Descriptor: | 1-ACETYL-4-(4-{[(2S,4R)-2-(2,4-DICHLOROPHENYL)-2-(1H-IMIDAZOL-1-YLMETHYL)-1,3-DIOXOLAN-4-YL]METHOXY}PHENYL)PIPERAZINE, CYTOCHROME P450 113A1, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Savino, C, Sciara, G, Miele, A.E, Kendrew, S.G, Vallone, B. | | Deposit date: | 2008-04-15 | | Release date: | 2009-07-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Azole Drugs Trap Cytochrome P450 Eryk in Alternative Conformational States.
Biochemistry, 49, 2010
|
|
2JFN
 
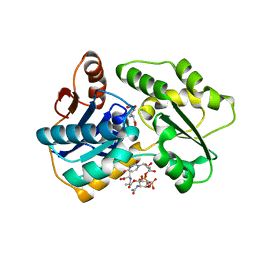 | |
2JFX
 
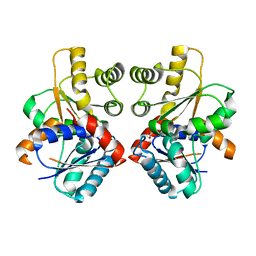 | |
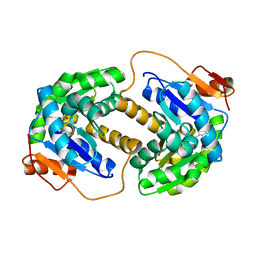
2JFO
 
 | |
2L3U
 
 | | Solution Structure of Domain IV from the YbbR family protein of Desulfitobacterium hafniense: Northeast Structural Genomics Consortium target DhR29A | | Descriptor: | YbbR family protein | | Authors: | Barb, A.W, Lee, H, Belote, R.L, Ciccosanti, C, Hamilton, K, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Prestegard, J.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-09-23 | | Release date: | 2010-10-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures of domains I and IV from YbbR are representative of a widely distributed protein family.
Protein Sci., 20, 2011
|
|
2LQL
 
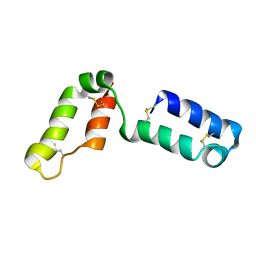 | | Solution structure of CHCH5 | | Descriptor: | Coiled-coil-helix-coiled-coil-helix domain-containing protein 5 | | Authors: | Peruzzini, R, Ciofi-Baffoni, S, Banci, L, Bertini, I. | | Deposit date: | 2012-03-09 | | Release date: | 2012-10-24 | | Last modified: | 2023-12-06 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of CHCHD5 and CHCHD7: Two atypical human twin CX(9)C proteins.
J.Struct.Biol., 180, 2012
|
|
2LQT
 
 | |
2LQ8
 
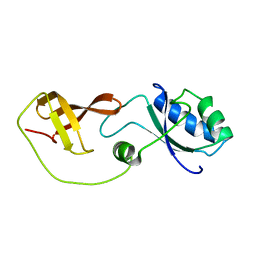 | | Domain interaction in Thermotoga maritima NusG | | Descriptor: | Transcription antitermination protein nusG | | Authors: | Droegemueller, J, Stegmann, C, Burmann, B, Roesch, P, Wahl, M.C, Schweimer, K. | | Deposit date: | 2012-02-27 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An Autoinhibited State in the Structure of Thermotoga maritima NusG.
Structure, 21, 2013
|
|
2LDO
 
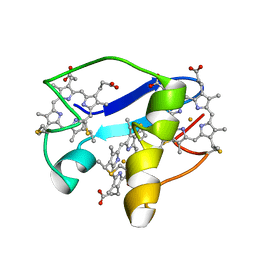 | | Solution structure of triheme cytochrome PpcA from Geobacter sulfurreducens reveals the structural origin of the redox-Bohr effect | | Descriptor: | Cytochrome c3, HEME C | | Authors: | Morgado, L, Paixao, V.B, Bruix, M, Salgueiro, C.A. | | Deposit date: | 2011-05-30 | | Release date: | 2011-09-07 | | Last modified: | 2021-03-03 | | Method: | SOLUTION NMR | | Cite: | Revealing the structural origin of the redox-Bohr effect: the first solution structure of a cytochrome from Geobacter sulfurreducens.
Biochem.J., 441, 2012
|
|
4K72
 
 | |
4C6J
 
 | | Crystal structure of the dihydroorotase domain of human CAD bound to substrate at pH 7.5 | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CAD PROTEIN, FORMIC ACID, ... | | Authors: | Ramon-Maiques, S, Lallous, N, Grande-Garcia, A. | | Deposit date: | 2013-09-18 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.299 Å) | | Cite: | Structure, Functional Characterization and Evolution of the Dihydroorotase Domain of Human Cad.
Structure, 22, 2014
|
|
4C6F
 
 | | Crystal structure of the dihydroorotase domain of human CAD bound to substrate at pH 6.5 | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, DIHYDROOROTASE, FORMIC ACID, ... | | Authors: | Ramon-Maiques, S, Lallous, N, Grande-Garcia, A. | | Deposit date: | 2013-09-18 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.261 Å) | | Cite: | Structure, Functional Characterization and Evolution of the Dihydroorotase Domain of Human Cad.
Structure, 22, 2014
|
|
