1J9Y
 
 | | Crystal structure of mannanase 26A from Pseudomonas cellulosa | | Descriptor: | MANNANASE A, ZINC ION | | Authors: | Hogg, D, Woo, E.-J, Bolam, D.N, McKie, V.A, Gilbert, H.J, Pickersgill, R.W. | | Deposit date: | 2001-05-29 | | Release date: | 2001-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of mannanase 26A from Pseudomonas cellulosa and analysis of residues involved in substrate binding
J.Biol.Chem., 276, 2001
|
|
5WHQ
 
 | | Crystal structure of the catalase-peroxidase from Neurospora crassa at 2.9 A | | Descriptor: | Catalase-peroxidase, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Diaz-Vilchis, A, Vega-Garcia, V, Rudino-Pinera, E, Hansberg, W. | | Deposit date: | 2017-07-18 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure, kinetics, molecular and redox properties of a cytosolic and developmentally regulated fungal catalase-peroxidase.
Arch. Biochem. Biophys., 640, 2018
|
|
3TZE
 
 | |
5YQW
 
 | | Structure and function of a novel periplasmic chitooligosaccharide-binding protein from marine Vibrio bacteria | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NICKEL (II) ION, ... | | Authors: | Suginta, W, Sritho, N, Ranok, A, Kitaoku, Y, Bulmer, D.M, van den Berg, B, Fukamizo, T. | | Deposit date: | 2017-11-08 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structure and function of a novel periplasmic chitooligosaccharide-binding protein from marineVibriobacteria.
J. Biol. Chem., 293, 2018
|
|
5YTU
 
 | |
5YPF
 
 | | p62/SQSTM1 ZZ domain with Trp-peptide | | Descriptor: | 78 kDa glucose-regulated protein,Sequestosome-1, ZINC ION | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-11-01 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter.
Nat Commun, 9, 2018
|
|
5YTO
 
 | |
7P1C
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with darobactin B | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Outer membrane protein assembly factor BamA, TRP-ASN-UX8-THR-LYS-ARG-PHE | | Authors: | Jakob, R.P, Modaresi, S.M, Hiller, S, Maier, T. | | Deposit date: | 2021-07-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutasynthetic Production and Antimicrobial Characterization of Darobactin Analogs.
Microbiol Spectr, 9, 2021
|
|
5YUL
 
 | | Native Structure of hSOD1 in P6322 space group | | Descriptor: | Dihydrogen tetrasulfide, GLYCEROL, Superoxide dismutase [Cu-Zn], ... | | Authors: | Manjula, R, Padmanabhan, B. | | Deposit date: | 2017-11-22 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Assessment of ligand binding at a site relevant to SOD1 oxidation and aggregation
FEBS Lett., 592, 2018
|
|
5L05
 
 | |
5WN8
 
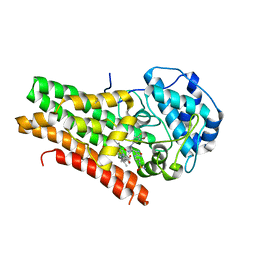 | | Structural Insights into Substrate and Inhibitor Binding Sites in Human Indoleamine 2,3-Dioxygenase 1 | | Descriptor: | Indoleamine 2,3-dioxygenase 1, N-(3-bromo-4-fluorophenyl)-N'-hydroxy-4-{[2-(sulfamoylamino)ethyl]amino}-1,2,5-oxadiazole-3-carboximidamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lewis-Ballester, A, Pham, K.N, Batabyal, D, Karkashon, S, Bonanno, J.B, Poulos, T.L, Yeh, S.R. | | Deposit date: | 2017-07-31 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into substrate and inhibitor binding sites in human indoleamine 2,3-dioxygenase 1.
Nat Commun, 8, 2017
|
|
5ZJ4
 
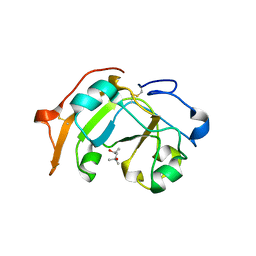 | | Guanine-specific ADP-ribosyltransferase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADP-ribosyltransferase | | Authors: | Yoshida, T, Tsuge, H. | | Deposit date: | 2018-03-19 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.49739277 Å) | | Cite: | Substrate N2atom recognition mechanism in pierisin family DNA-targeting, guanine-specific ADP-ribosyltransferase ScARP.
J. Biol. Chem., 293, 2018
|
|
5ZJ5
 
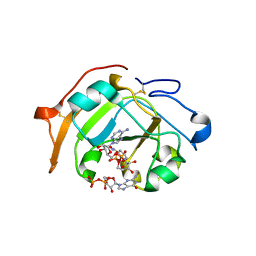 | | Guanine-specific ADP-ribosyltransferase with NADH and GDP | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ADP-ribosyltransferase, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Yoshida, T, Tsuge, H. | | Deposit date: | 2018-03-19 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.568112 Å) | | Cite: | Substrate N2atom recognition mechanism in pierisin family DNA-targeting, guanine-specific ADP-ribosyltransferase ScARP.
J. Biol. Chem., 293, 2018
|
|
5WMW
 
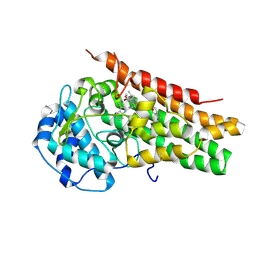 | | Structural Insights into Substrate and Inhibitor Binding Sites in Human Indoleamine 2,3-Dioxygenase 1 | | Descriptor: | CYANIDE ION, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lewis-Ballester, A, Yeh, S.R, Pham, K.N, Batabyal, D, Karkashon, S, Bonanno, J.B, Poulos, T.L. | | Deposit date: | 2017-07-31 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Structural insights into substrate and inhibitor binding sites in human indoleamine 2,3-dioxygenase 1.
Nat Commun, 8, 2017
|
|
6LW5
 
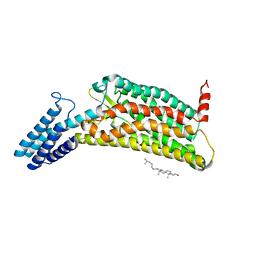 | | Crystal structure of the human formyl peptide receptor 2 in complex with WKYMVm | | Descriptor: | CHOLESTEROL, Soluble cytochrome b562,N-formyl peptide receptor 2, TRP-LYS-TYR-MET-VAL-QXV | | Authors: | Chen, T, Zong, X, Zhang, H, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2020-02-07 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of ligand binding modes at the human formyl peptide receptor 2.
Nat Commun, 11, 2020
|
|
4WYU
 
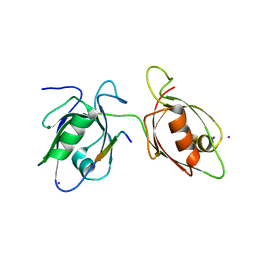 | |
4WK4
 
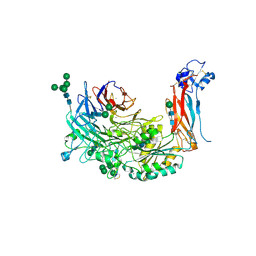 | | Metal Ion and Ligand Binding of Integrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ALA-CYS-ARG-GLY-ASP-GLY-TRP-CYS, ... | | Authors: | Xia, W, Springer, T.A. | | Deposit date: | 2014-10-01 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Metal ion and ligand binding of integrin alpha 5 beta 1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4OAA
 
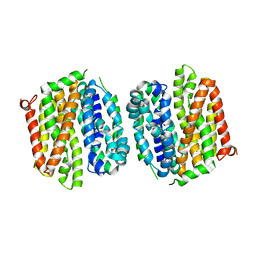 | | Crystal structure of E. coli lactose permease G46W,G262W bound to sugar | | Descriptor: | Lactose/galactose transporter, beta-D-galactopyranose-(1-1)-1-thio-beta-D-galactopyranose | | Authors: | Kumar, H, Kasho, V, Smirnova, I, Finer-Moore, J, Kaback, H.R, Stroud, R.M. | | Deposit date: | 2014-01-03 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of sugar-bound LacY.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1CCK
 
 | | ALTERING SUBSTRATE SPECIFICITY OF CYTOCHROME C PEROXIDASE TOWARDS A SMALL MOLECULAR SUBSTRATE PEROXIDASE BY SUBSTITUTING TYROSINE FOR PHE 202 | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Cao, Y, Musah, R.A, Wilcox, S.K, Goodin, D.B, Mcree, D.E. | | Deposit date: | 1998-01-02 | | Release date: | 1998-11-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of recombinant pea cytosolic ascorbate peroxidase.
Biochemistry, 34, 1995
|
|
1CCP
 
 | | X-RAY STRUCTURES OF RECOMBINANT YEAST CYTOCHROME C PEROXIDASE AND THREE HEME-CLEFT MUTANTS PREPARED BY SITE-DIRECTED MUTAGENESIS | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, YEAST CYTOCHROME C PEROXIDASE | | Authors: | Wang, J, Mauro, J.M, Edwards, S.L, Oatley, S.J, Fishel, L.A, Ashford, V.A, Xuong, N.-H, Kraut, J. | | Deposit date: | 1990-02-28 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structures of recombinant yeast cytochrome c peroxidase and three heme-cleft mutants prepared by site-directed mutagenesis.
Biochemistry, 29, 1990
|
|
1TP5
 
 | | Crystal structure of PDZ3 domain of PSD-95 protein complexed with a peptide ligand KKETWV | | Descriptor: | LYS-LYS-GLU-THR-TRP-VAL peptide ligand, Presynaptic density protein 95 | | Authors: | Saro, D, Wawrzak, Z, Martin, P, Vickrey, J, Paredes, A, Kovari, L, Spaller, M. | | Deposit date: | 2004-06-15 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure of the third PDZ domain of PSD-95 protein complexed with KKETWV peptide ligand
To be Published
|
|
2E4G
 
 | | RebH with bound L-Trp | | Descriptor: | TRYPTOPHAN, Tryptophan halogenase | | Authors: | Blasiak, L.C, Drennan, C.L. | | Deposit date: | 2006-12-07 | | Release date: | 2007-02-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Chlorination by a long-lived intermediate in the mechanism of flavin-dependent halogenases
Biochemistry, 46, 2007
|
|
1WDW
 
 | | Structural basis of mutual activation of the tryptophan synthase a2b2 complex from a hyperthermophile, Pyrococcus furiosus | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Tryptophan synthase alpha chain, Tryptophan synthase beta chain 1 | | Authors: | Lee, S.J, Ogasahara, K, Ma, J, Nishio, K, Ishida, M, Yamagata, Y, Tsukihara, T, Yutani, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-19 | | Release date: | 2005-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Conformational Changes in the Tryptophan Synthase from a Hyperthermophile upon alpha(2)beta(2) Complex Formation: Crystal Structure of the Complex
Biochemistry, 44, 2005
|
|
4ICS
 
 | | Crystal structure of PepS from Streptococcus pneumoniae in complex with a substrate | | Descriptor: | Aminopeptidase PepS, GLYCINE, TRYPTOPHAN, ... | | Authors: | Lee, S, Kim, K.K, Ta, M.H. | | Deposit date: | 2012-12-11 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure-based elucidation of the regulatory mechanism for aminopeptidase activity.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
8ADI
 
 | | Cryo-EM structure of Darobactin 9 bound BAM complex | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Yuan, B, Marlovits, T.C. | | Deposit date: | 2022-07-08 | | Release date: | 2023-01-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Darobactins Exhibiting Superior Antibiotic Activity by Cryo-EM Structure Guided Biosynthetic Engineering.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
