6VH0
 
 | | 1.95 A resolution structure of MERS 3CL protease in complex with inhibitor 6g | | Descriptor: | N~2~-{[(5-ethyl-1,3-dioxan-5-yl)oxy]carbonyl}-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide, Orf1a protein | | Authors: | Lovell, S, Battaile, K.P, Kashipathy, M.M, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-01-09 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
6VH1
 
 | | 2.30 A resolution structure of MERS 3CL protease in complex with inhibitor 6h | | Descriptor: | N~2~-{[(4,4-difluorocyclohexyl)oxy]carbonyl}-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide, Orf1a protein | | Authors: | Lovell, S, Battaile, K.P, Kashipathy, M.M, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-01-09 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
6VGZ
 
 | | 2.25 A resolution structure of MERS 3CL protease in complex with inhibitor 6d | | Descriptor: | N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-({[trans-4-(propan-2-yl)cyclohexyl]oxy}carbonyl)-L-leucinamide, Orf1a protein | | Authors: | Lovell, S, Battaile, K.P, Kashipathy, M.M, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-01-09 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
6SD9
 
 | | Crystal structure of wild-type cMET bound by foretinib | | Descriptor: | CHLORIDE ION, Hepatocyte growth factor receptor, N-(3-fluoro-4-{[6-methoxy-7-(3-morpholin-4-ylpropoxy)quinolin-4-yl]oxy}phenyl)-N'-(4-fluorophenyl)cyclopropane-1,1-dicarboxamide | | Authors: | Collie, G.W, Phillips, C. | | Deposit date: | 2019-07-26 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and Molecular Insight into Resistance Mechanisms of First Generation cMET Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6SDC
 
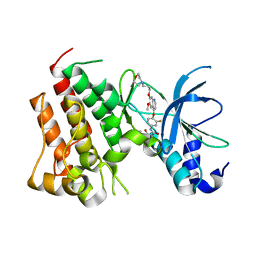 | | Crystal structure of D1228V cMET bound by foretinib | | Descriptor: | Hepatocyte growth factor receptor, N-(3-fluoro-4-{[6-methoxy-7-(3-morpholin-4-ylpropoxy)quinolin-4-yl]oxy}phenyl)-N'-(4-fluorophenyl)cyclopropane-1,1-dicarboxamide | | Authors: | Collie, G.W, Phillips, C. | | Deposit date: | 2019-07-26 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural and Molecular Insight into Resistance Mechanisms of First Generation cMET Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6SDD
 
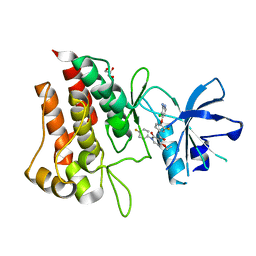 | | Crystal structure of D1228V cMET bound by BMS-777607 | | Descriptor: | GLYCEROL, Hepatocyte growth factor receptor, N-{4-[(2-amino-3-chloropyridin-4-yl)oxy]-3-fluorophenyl}-4-ethoxy-1-(4-fluorophenyl)-2-oxo-1,2-dihydropyridine-3-carboxamide | | Authors: | Collie, G.W, Phillips, C. | | Deposit date: | 2019-07-26 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural and Molecular Insight into Resistance Mechanisms of First Generation cMET Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6SDE
 
 | |
8SFE
 
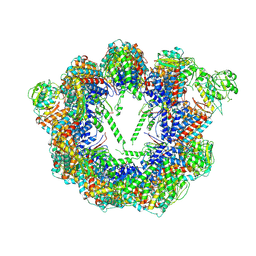 | | Open state CCT-G beta 5 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
7BF3
 
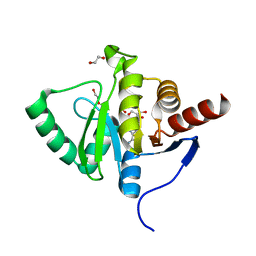 | | Crystal structure of SARS-CoV-2 macrodomain in complex with adenosine | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE, MAGNESIUM ION, ... | | Authors: | Ni, X, Knapp, S, Chaikuad, A, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-31 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
7BF6
 
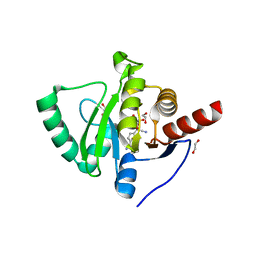 | | Crystal structure of SARS-CoV-2 macrodomain in complex with remdesivir metabolite GS-441524 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(4-azanylpyrrolo[2,1-f][1,2,4]triazin-7-yl)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolane-2-carbonitrile, 1,2-ETHANEDIOL, Papain-like protease nsp3 | | Authors: | Ni, X, Knapp, S, Chaikuad, A, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-31 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
6NPM
 
 | |
6XF8
 
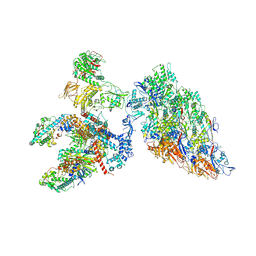 | | DLP 5 fold | | Descriptor: | Inner capsid protein lambda-1, Inner capsid protein sigma-2, Outer capsid protein mu-1, ... | | Authors: | Sutton, G, Sun, D.P, Fu, X.F, Kotecha, A, Hecksel, G.W, Clare, D.K, Zhang, P, Stuart, D, Boyce, M. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Assembly intermediates of orthoreovirus captured in the cell.
Nat Commun, 11, 2020
|
|
7BF4
 
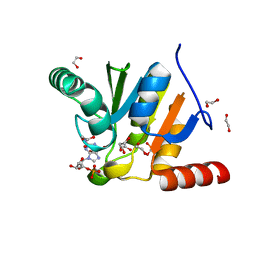 | | Crystal structure of SARS-CoV-2 macrodomain in complex with GMP | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-MONOPHOSPHATE, NSP3 macrodomain | | Authors: | Ni, X, Knapp, S, Chaikuad, A, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-31 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
6NPI
 
 | | Crystal structure of Epstein-Barr Virus Nuclear Antigen-1, EBNA1, bound to fragments | | Descriptor: | ({2-[(4-bromo-5-methyl-1,2-oxazol-3-yl)amino]-2-oxoethyl}sulfanyl)acetic acid, 2-pyrrol-1-ylbenzoic acid, Epstein-Barr nuclear antigen 1 | | Authors: | Messick, T.E. | | Deposit date: | 2019-01-17 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Structure-based design of small-molecule inhibitors of EBNA1 DNA binding blocks Epstein-Barr virus latent infection and tumor growth.
Sci Transl Med, 11, 2019
|
|
6NX1
 
 | | STRUCTURE OF HUMAN PREGNANE X RECEPTOR LIGAND BINDING DOMAIN BOUND TETHERED WITH SRC CO-ACTIVATOR PEPTIDE AND COMPOUND-3 AKA 1,1,1,3,3,3-HEXAFLUORO-2-{4-[1-(4- LUOROBENZENESULFONYL)CYCLOPENTYL]PHENYL}PROPAN-2-OL | | Descriptor: | 1,1,1,3,3,3-hexafluoro-2-(4-{1-[(4-fluorophenyl)sulfonyl]cyclopentyl}phenyl)propan-2-ol, Nuclear receptor subfamily 1 group I member 2,Nuclear receptor coactivator 1 fusion | | Authors: | Khan, J.A. | | Deposit date: | 2019-02-07 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure-based Discovery of Phenyl (3-Phenylpyrrolidin-3-yl)sulfones as Selective, Orally Active ROR gamma t Inverse Agonists.
Acs Med.Chem.Lett., 10, 2019
|
|
6NY4
 
 | |
6S1I
 
 | | Crystal Structure of DYRK1A with small molecule inhibitor | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 1A, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Sorrell, F.J, Henderson, S.H, Redondo, C, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Elkins, J.M. | | Deposit date: | 2019-06-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Mining Public Domain Data to Develop Selective DYRK1A Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
7BF5
 
 | | Crystal structure of SARS-CoV-2 macrodomain in complex with ADP-ribose-phosphate (ADP-ribose-2'-phosphate, ADPRP) | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, NSP3 macrodomain, ... | | Authors: | Ni, X, Knapp, S, Chaikuad, A, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-31 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
6NPP
 
 | |
8AX5
 
 | | Crystal structure of a CGRP receptor ectodomain heterodimer bound to macrocyclic inhibitor HTL0029881 | | Descriptor: | (1~{R},10~{R},20~{E})-12-methyl-10-[(7-methyl-2~{H}-indazol-5-yl)methyl]-15,18-dioxa-9,12,24,26-tetrazapentacyclo[20.5.2.1^{1,4}.1^{3,7}.0^{25,28}]hentriaconta-3,5,7(30),20,22,24,28-heptaene-8,11,27-trione, Maltose/maltodextrin-binding periplasmic protein,Receptor activity-modifying protein 1,Calcitonin gene-related peptide type 1 receptor, TETRAETHYLENE GLYCOL, ... | | Authors: | Southall, S.M, Watson, S.P. | | Deposit date: | 2022-08-30 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Novel Macrocyclic Antagonists of the CGRP Receptor Part 2: Stereochemical Inversion Induces an Unprecedented Binding Mode.
Acs Med.Chem.Lett., 13, 2022
|
|
8AX6
 
 | | Crystal structure of a CGRP receptor ectodomain heterodimer bound to macrocyclic inhibitor HTL0029882 | | Descriptor: | (1~{S},10~{R},20~{E})-12-methyl-10-[(7-methyl-2~{H}-indazol-5-yl)methyl]-15,18-dioxa-9,12,24,26-tetrazapentacyclo[20.5.2.1^{1,4}.1^{3,7}.0^{25,28}]hentriaconta-3(30),4,6,20,22,24,28-heptaene-8,11,27-trione, Maltose/maltodextrin-binding periplasmic protein,Receptor activity-modifying protein 1,Calcitonin gene-related peptide type 1 receptor, TETRAETHYLENE GLYCOL, ... | | Authors: | Southall, S.M, Watson, S.P. | | Deposit date: | 2022-08-30 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel Macrocyclic Antagonists of the CGRP Receptor Part 2: Stereochemical Inversion Induces an Unprecedented Binding Mode.
Acs Med.Chem.Lett., 13, 2022
|
|
8AX7
 
 | | Crystal structure of a CGRP receptor ectodomain heterodimer bound to macrocyclic inhibitor HTL0031448 | | Descriptor: | (1~{S},10~{R},20~{E})-10-[(1,7-dimethylindazol-5-yl)methyl]-12-methyl-15,18-dioxa-9,12,24,26-tetrazapentacyclo[20.5.2.1^{1,4}.1^{3,7}.0^{25,28}]hentriaconta-3(30),4,6,20,22,24,28-heptaene-8,11,27-trione, ACETATE ION, Maltose/maltodextrin-binding periplasmic protein,Receptor activity-modifying protein 1,Calcitonin gene-related peptide type 1 receptor, ... | | Authors: | Southall, S.M, Watson, S.P. | | Deposit date: | 2022-08-30 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Novel Macrocyclic Antagonists of the CGRP Receptor Part 2: Stereochemical Inversion Induces an Unprecedented Binding Mode.
Acs Med.Chem.Lett., 13, 2022
|
|
5BOY
 
 | |
5ULN
 
 | | Synthesis of novel seleno ureido containing compounds as SLC-0111 analogs. Investigations on carbonic anhydrases activity, glutathione peroxidase and X-ray crystallography | | Descriptor: | 4-{[(4-fluorophenyl)carbamothioyl]amino}benzene-1-sulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Peat, T.S, Angeli, A, Tanini, D, Bartolucci, G, Capperucci, A, Supuran, C.T, Carta, F. | | Deposit date: | 2017-01-25 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Discovery of New Selenoureido Analogues of 4-(4-Fluorophenylureido)benzenesulfonamide as Carbonic Anhydrase Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
5UMC
 
 | | Synthesis of novel seleno ureido containing compounds as SLC-0111 analogs. Investigations on carbonic anhydrases activity, glutathione peroxidase and X-ray crystallography | | Descriptor: | Carbonic anhydrase 2, GLYCEROL, UNKNOWN LIGAND, ... | | Authors: | Peat, T.S, Angeli, A, Tanini, D, Bartolucci, G, Capperucci, A, Supuran, C.T, Carta, F. | | Deposit date: | 2017-01-26 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of New Selenoureido Analogues of 4-(4-Fluorophenylureido)benzenesulfonamide as Carbonic Anhydrase Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
