7NWG
 
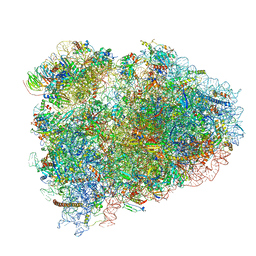 | | Mammalian pre-termination 80S ribosome with Hybrid P/E- and A/P-site tRNA's bound by Blasticidin S. | | Descriptor: | 18S Ribosomal RNA, 28S Ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Powers, K.T, Yadav, S.K.N, Bufton, J.C, Schaffitzel, C. | | Deposit date: | 2021-03-16 | | Release date: | 2021-07-07 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Blasticidin S inhibits mammalian translation and enhances production of protein encoded by nonsense mRNA.
Nucleic Acids Res., 49, 2021
|
|
7NWH
 
 | | Mammalian pre-termination 80S ribosome with eRF1 and eRF3 bound by Blasticidin S. | | Descriptor: | 18S Ribosomal RNA, 28S Ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Powers, K.T, Yadav, S.K.N, Bufton, J.C, Schaffitzel, C. | | Deposit date: | 2021-03-16 | | Release date: | 2021-07-07 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Blasticidin S inhibits mammalian translation and enhances production of protein encoded by nonsense mRNA.
Nucleic Acids Res., 49, 2021
|
|
9GR0
 
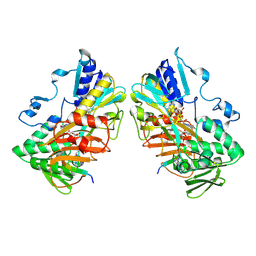 | | Interaction with AK2A links AIFM1 to cellular energy metabolism. The cryo-EM structure of dimeric AIFM1 bound by AK2A. | | Descriptor: | Adenylate kinase 2, mitochondrial, Apoptosis-inducing factor 1, ... | | Authors: | Rothemann, R.A, Pavlenko, E.A, Gerlich, S, Grobushkin, P, Mostert, S, Stobbe, D, Racho, J, Stillger, K, Lapacz, K, Petrungaro, C, Dengjel, J, Neundorf, I, Bano, D, Mondal, M, Weiss, K, Ehninger, D, Nguyen, T.H.D, Poepsel, S.P, Riemer, J. | | Deposit date: | 2024-09-10 | | Release date: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Interaction with AK2A links AIFM1 to cellular energy metabolism.
Mol.Cell, 2025
|
|
6DQB
 
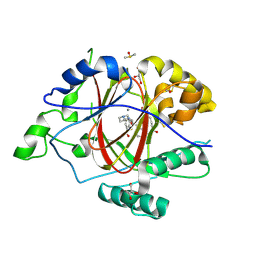 | | LINKED KDM5A JMJ DOMAIN FORMING COVALENT BOND TO INHIBITOR N71 i.e. 2-((3-(4-(dimethylamino)but-2-enamido)phenyl)(2-(piperidin-1-yl)ethoxy)methyl)thieno[3,2-b]pyridine-7-carboxylic acid | | Descriptor: | 2-{(R)-(3-{[(2E)-4-(dimethylamino)but-2-enoyl]amino}phenyl)[2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, 2-{(R)-(3-{[4-(dimethylamino)butanoyl]amino}phenyl)[2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, 2-{(S)-(3-{[4-(dimethylamino)butanoyl]amino}phenyl)[2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-06-10 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
6DQ6
 
 | |
6DQF
 
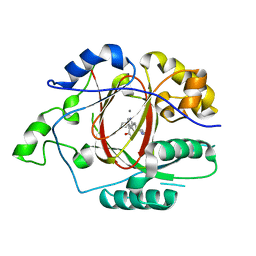 | |
7ODC
 
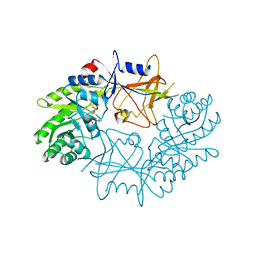 | | CRYSTAL STRUCTURE ORNITHINE DECARBOXYLASE FROM MOUSE, TRUNCATED 37 RESIDUES FROM THE C-TERMINUS, TO 1.6 ANGSTROM RESOLUTION | | Descriptor: | PROTEIN (ORNITHINE DECARBOXYLASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Kern, A.D, Oliveira, M.A, Coffino, P, Hackert, M.L. | | Deposit date: | 1999-03-03 | | Release date: | 1999-10-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of mammalian ornithine decarboxylase at 1.6 A resolution: stereochemical implications of PLP-dependent amino acid decarboxylases.
Structure Fold.Des., 7, 1999
|
|
10MH
 
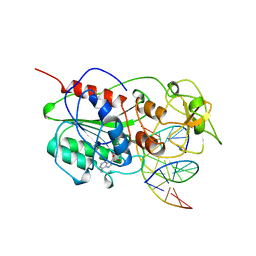 | | TERNARY STRUCTURE OF HHAI METHYLTRANSFERASE WITH ADOHCY AND HEMIMETHYLATED DNA CONTAINING 5,6-DIHYDRO-5-AZACYTOSINE AT THE TARGET | | Descriptor: | DNA (5'-D(P*CP*CP*AP*TP*GP*(5CM)P*GP*CP*TP*GP*AP*C)-3'), DNA (5'-D(P*GP*TP*CP*AP*GP*5NCP*GP*CP*AP*TP*GP*G)-3'), PROTEIN (CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI), ... | | Authors: | Sheikhnejad, G, Brank, A, Christman, J.K, Goddard, A, Alvarez, E, Ford Junior, H, Marquez, V.E, Marasco, C.J, Sufrin, J.R, O'Gara, M, Cheng, X. | | Deposit date: | 1998-08-10 | | Release date: | 1999-02-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Mechanism of inhibition of DNA (cytosine C5)-methyltransferases by oligodeoxyribonucleotides containing 5,6-dihydro-5-azacytosine.
J.Mol.Biol., 285, 1999
|
|
6DQA
 
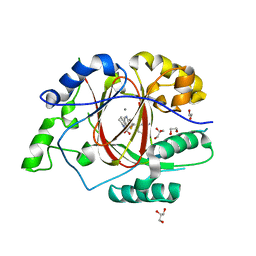 | | Linked KDM5A JMJ Domain Bound to Inhibitor N70 i.e.[2-((3-aminophenyl)(2-(piperidin-1-yl)ethoxy)methyl)thieno[3,2-b]pyridine-7-carboxylic acid] | | Descriptor: | 1,2-ETHANEDIOL, 2-{(R)-(3-aminophenyl)[2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-06-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.888 Å) | | Cite: | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
6BGX
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)((4,4-difluorocyclohexyl)methoxy)methyl)-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid(Compound N42) | | Descriptor: | 1,2-ETHANEDIOL, 2-{(S)-(2-chlorophenyl)[(4,4-difluorocyclohexyl)methoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
1AKE
 
 | |
1AK2
 
 | | ADENYLATE KINASE ISOENZYME-2 | | Descriptor: | ADENYLATE KINASE ISOENZYME-2, SULFATE ION | | Authors: | Schlauderer, G.J, Schulz, G.E. | | Deposit date: | 1995-12-29 | | Release date: | 1996-06-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The structure of bovine mitochondrial adenylate kinase: comparison with isoenzymes in other compartments.
Protein Sci., 5, 1996
|
|
7OSA
 
 | | Pre-translocation complex of 80 S.cerevisiae ribosome with eEF2 and ligands | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0, ... | | Authors: | Djumagulov, M, Jenner, L, Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2021-06-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Accuracy mechanism of eukaryotic ribosome translocation.
Nature, 600, 2021
|
|
7OSM
 
 | | Intermediate translocation complex of 80 S.cerevisiae ribosome with eEF2 and ligands | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0, ... | | Authors: | Djumagulov, M, Jenner, L, Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2021-06-09 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Accuracy mechanism of eukaryotic ribosome translocation.
Nature, 600, 2021
|
|
7OLD
 
 | | Thermophilic eukaryotic 80S ribosome at pe/E (TI)-POST state | | Descriptor: | 18S rRNA, 26S rRNA, 40S ribosomal protein S0, ... | | Authors: | Kisonaite, M, Wild, K, Sinning, I. | | Deposit date: | 2021-05-19 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | High-resolution structures of a thermophilic eukaryotic 80S ribosome reveal atomistic details of translocation.
Nat Commun, 13, 2022
|
|
7OLC
 
 | | Thermophilic eukaryotic 80S ribosome at idle POST state | | Descriptor: | 18S rRNA, 26S rRNA, 40S ribosomal protein S0, ... | | Authors: | Kisonaite, M, Wild, K, Sinning, I. | | Deposit date: | 2021-05-19 | | Release date: | 2022-01-26 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | High-resolution structures of a thermophilic eukaryotic 80S ribosome reveal atomistic details of translocation.
Nat Commun, 13, 2022
|
|
6BGZ
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)(2-(1-methyl-1H-imidazol-2-yl)ethoxy)methyl)-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid (Compound N47) | | Descriptor: | 2-{(S)-(2-chlorophenyl)[2-(1-methyl-1H-imidazol-2-yl)ethoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, GLYCEROL, Lysine-specific demethylase 5A, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.688 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6BH1
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR (S)-2-((2-chlorophenyl)(2-(piperidin-1-yl)ethoxy)methyl)-1l2-pyrrolo[3,2-b]pyridine-7-carboxylic acid (Compound N52) | | Descriptor: | 1,2-ETHANEDIOL, 2-{(S)-(2-chlorophenyl)[2-(piperidin-1-yl)ethoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.932 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6DQ9
 
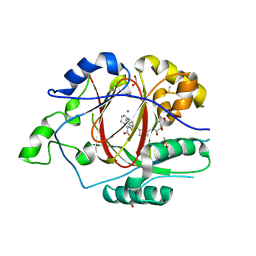 | | Linked KDM5A JMJ Domain Bound to the Covalent Inhibitor N69 i.e. [2-((3-acrylamidophenyl)(2-(piperidin-1-yl)ethoxy)methyl)thieno[3,2-b]pyridine-7-carboxylic acid] | | Descriptor: | 1,2-ETHANEDIOL, 2-{(R)-[3-(acryloylamino)phenyl][2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-06-10 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.748 Å) | | Cite: | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
1ANK
 
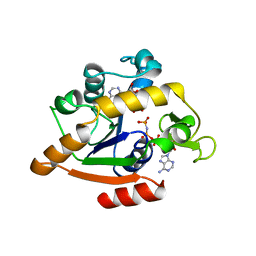 | | THE CLOSED CONFORMATION OF A HIGHLY FLEXIBLE PROTEIN: THE STRUCTURE OF E. COLI ADENYLATE KINASE WITH BOUND AMP AND AMPPNP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENYLATE KINASE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Berry, M.B, Meador, B, Bilderback, T, Liang, P, Glaser, M, Phillips Jr, G.N. | | Deposit date: | 1994-02-28 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The closed conformation of a highly flexible protein: the structure of E. coli adenylate kinase with bound AMP and AMPPNP.
Proteins, 19, 1994
|
|
7E9V
 
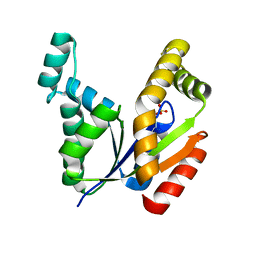 | | The Crystal Structure of human UMP-CMP kinase from Biortus. | | Descriptor: | SULFATE ION, UMP-CMP kinase | | Authors: | Wang, F, Lin, D, Wang, R, Wei, X, Shen, Z, Wang, M. | | Deposit date: | 2021-03-05 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of human UMP-CMP kinase from Biortus.
To Be Published
|
|
6DQ5
 
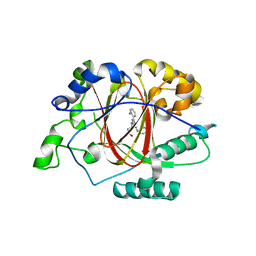 | |
6DQE
 
 | |
7OYA
 
 | | Cryo-EM structure of the 1 hpf zebrafish embryo 80S ribosome | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Leesch, F, Lorenzo-Orts, L, Grishkovskaya, I, Kandolf, S, Belacic, K, Meinhart, A, Haselbach, D, Pauli, A. | | Deposit date: | 2021-06-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A molecular network of conserved factors keeps ribosomes dormant in the egg.
Nature, 613, 2023
|
|
7OYD
 
 | | Cryo-EM structure of a rabbit 80S ribosome with zebrafish Dap1b | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Leesch, F, Lorenzo-Orts, L, Grishkovskaya, I, Kandolf, S, Belacic, K, Meinhart, A, Haselbach, D, Pauli, A. | | Deposit date: | 2021-06-24 | | Release date: | 2022-07-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | A molecular network of conserved factors keeps ribosomes dormant in the egg.
Nature, 613, 2023
|
|
