8PXL
 
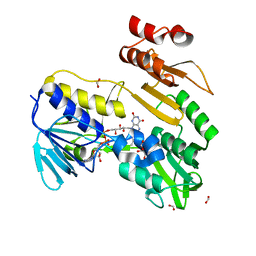 | | Structure of NADH-DEPENDENT FERREDOXIN REDUCTASE, BPHA4, solved at wavelength 1.37 A | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, Ferredoxin reductase, ... | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Senda, M, Matsugaki, N, Kawano, Y, Wagner, A. | | Deposit date: | 2023-07-23 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8SXQ
 
 | |
7UCW
 
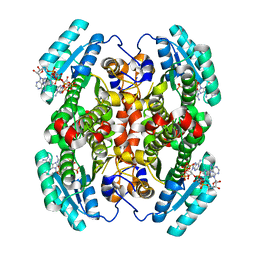 | | Structure of mouse Decr1 in complex with 2'-5' oligoadenylate | | Descriptor: | Decr1 protein, [[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-4-[[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-4-[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-3-hydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-3-hydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl] phosphono hydrogen phosphate | | Authors: | Govande, A.A, Kranzusch, P.J. | | Deposit date: | 2022-03-17 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | RNase L-activating 2'-5' oligoadenylates bind ABCF1, ABCF3 and Decr-1.
J.Gen.Virol., 104, 2023
|
|
7PR7
 
 | | Crystal structure of human heparanase in complex with covalent inhibitor VL166 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-(2R,3S,5R,6R)-2,3,4,5,6-pentakis(oxidanyl)cyclohexane-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, L, Armstrong, Z, Davies, G.J. | | Deposit date: | 2021-09-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Mechanism-based heparanase inhibitors reduce cancer metastasis in vivo.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8PXH
 
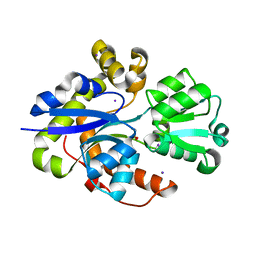 | | Structure of TauA from E. coli, solved at wavelength 2.375 A | | Descriptor: | 2-AMINOETHANESULFONIC ACID, IODIDE ION, Taurine ABC transporter substrate-binding protein | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Qu, F, Beis, K, Wagner, A. | | Deposit date: | 2023-07-23 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
7N8F
 
 | | The crystal structure of I38T mutant PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988288 | | Descriptor: | Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2021-06-14 | | Release date: | 2022-06-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
5DPR
 
 | |
6NSV
 
 | | Crystal structure of the human CHIP TPR domain in complex with a 5mer acetylated optimized peptide | | Descriptor: | ACE-LEU-TRP-TRP-PRO-ASP, CHLORIDE ION, E3 ubiquitin-protein ligase CHIP, ... | | Authors: | Basu, K, Ravalin, M, Bohn, M.-F, Craik, C.S, Gestwicki, J.E. | | Deposit date: | 2019-01-25 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.305 Å) | | Cite: | Specificity for latent C termini links the E3 ubiquitin ligase CHIP to caspases.
Nat.Chem.Biol., 15, 2019
|
|
7Z68
 
 | | Pseudomonas aeruginosa Elastase in complex with a Thiol based inhibitor (R-and S-configured) | | Descriptor: | (2~{R})-~{N}-[4-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]sulfanylphenyl]-3-phenyl-2-sulfanyl-propanamide, CALCIUM ION, GLYCEROL, ... | | Authors: | Klein, A, Hirsch, A. | | Deposit date: | 2022-03-11 | | Release date: | 2023-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Dual inhibitors of Pseudomonas aeruginosa virulence factors LecA and LasB
To Be Published
|
|
7N60
 
 | | The crystal structure of 4-methoxybenzoate-bound CYP199A S244D mutant | | Descriptor: | 4-METHOXYBENZOIC ACID, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Lau, I.C.K, Coleman, T, Bell, S.G, Bruning, J.B. | | Deposit date: | 2021-06-07 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | The crystal structure of 4-methoxybenzoate-bound CYP199A S244D mutant
To Be Published
|
|
8WG0
 
 | | Crystal structure of GH97 glucodextranase from Flavobacterium johnsoniae in complex with glucose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Candidate alpha-glucosidase Glycoside hydrolase family 97, ... | | Authors: | Kurata, R, Nakamura, S, Miyazaki, T. | | Deposit date: | 2023-09-20 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into alpha-(1→6)-linkage preference of GH97 glucodextranase from Flavobacterium johnsoniae.
Febs J., 291, 2024
|
|
6NTB
 
 | |
7Z8Z
 
 | | Crystal structure of the MEILB2-BRME1 2:2 core complex | | Descriptor: | Break repair meiotic recombinase recruitment factor 1, Heat shock factor 2-binding protein | | Authors: | Gurusaran, M, Davies, O.R. | | Deposit date: | 2022-03-19 | | Release date: | 2023-09-20 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | MEILB2-BRME1 forms a V-shaped DNA clamp upon BRCA2-binding in meiotic recombination.
Nat Commun, 15, 2024
|
|
5VDP
 
 | |
8WG1
 
 | | Crystal structure of GH97 glucodextranase mutant E509Q from Flavobacterium johnsoniae in complex with panose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Candidate alpha-glucosidase Glycoside hydrolase family 97, ... | | Authors: | Kurata, R, Nakamura, S, Miyazaki, T. | | Deposit date: | 2023-09-20 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural insights into alpha-(1→6)-linkage preference of GH97 glucodextranase from Flavobacterium johnsoniae.
Febs J., 291, 2024
|
|
7PR8
 
 | | Crystal structure of human heparanase in complex with covalent inhibitor GR109 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-(2R,3S,5R,6R)-2,3,4,5,6-pentakis(oxidanyl)cyclohexane-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, L, Armstrong, Z, Davies, G.J. | | Deposit date: | 2021-09-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Mechanism-based heparanase inhibitors reduce cancer metastasis in vivo.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7N8U
 
 | |
8WG2
 
 | | Crystal structure of GH97 glucodextranase mutant E509Q from Flavobacterium johnsoniae in complex with isomaltotriose | | Descriptor: | CALCIUM ION, Candidate alpha-glucosidase Glycoside hydrolase family 97, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose, ... | | Authors: | Kurata, R, Nakamura, S, Miyazaki, T. | | Deposit date: | 2023-09-20 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural insights into alpha-(1→6)-linkage preference of GH97 glucodextranase from Flavobacterium johnsoniae.
Febs J., 291, 2024
|
|
6NF3
 
 | |
7PZW
 
 | | LpxC Inhibitors With Fluoroproline As A Novel Zinc-Binding Group Can Serve As A Novel Class of Antibiotic With Activity Against Multidrug-Resistant Gram-Negative Bacteria | | Descriptor: | (4S)-N-((3R,5R)-1-(cyclopropanecarbonyl)-5-((5-(phenylethynyl)thiophene-2-carboxamido)methyl)pyrrolidin-3-yl)-4-fluoropyrrolidine-2-carboxamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Ryan, M.D, Pallin, T.D, Lamers, M.B.A.C, Leonard, P.M. | | Deposit date: | 2021-10-13 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | LpxC Inhibitors With Fluoroproline As A Novel Zinc-Binding Group Can Serve As A Novel Class of Antibiotic With Activity Against Multidrug-Resistant Gram-Negative Bacteria
To Be Published
|
|
8VF3
 
 | |
5DYX
 
 | |
7N9J
 
 | | Crystal structure of H2DB in complex with HSF2 melanoma neoantigen | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Patskovsky, Y, Finnigan, J, Patskovska, L, Newman, J, Bhardwaj, N, Krogsgaard, M. | | Deposit date: | 2021-06-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure of the complex between H2DB and melanoma HSF2 neoantigen YGFRNVVHI
To be Published
|
|
7NA5
 
 | | Structure of the H2DB-TCR ternary complex with HSF2 melanoma neoantigen | | Descriptor: | 47BE7 TCR alpha chain, 47BE7 TCR beta chain, Beta-2-microglobulin, ... | | Authors: | Patskovsky, Y, Finnigan, J, Patskovska, L, Newman, J, Bhardwaj, N, Krogsgaard, M. | | Deposit date: | 2021-06-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the TCR-H2DB ternary complex with melanoma HSF2 neoantigen YGFRNVVHI
To be Published
|
|
7PRT
 
 | | Crystal structure of human heparanase in complex with covalent inhibitor CB678 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-alpha-D-arabino-hexopyranose-(1-4)-(2R,3S,5R,6R)-2,3,4,5,6-pentakis(oxidanyl)cyclohexane-1-carboxylic acid, ... | | Authors: | Wu, L, Armstrong, Z, Davies, G.J. | | Deposit date: | 2021-09-22 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism-based heparanase inhibitors reduce cancer metastasis in vivo.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
