7O06
 
 | |
4FYJ
 
 | |
1PH2
 
 | |
8EEV
 
 | | Venezuelan equine encephalitis virus-like particle in complex with Fab SKT-20 | | Descriptor: | Coat protein, Fab SKT20 heavy chain, Fab SKT20 light chain | | Authors: | Tsybovsky, Y, Pletnev, S, Verardi, R, Roederer, M, Kwong, P.D. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Vaccine elicitation and structural basis for antibody protection against alphaviruses.
Cell, 186, 2023
|
|
5EAK
 
 | |
8F3C
 
 | | Cryo-EM consensus structure of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase minus preQ1 ligand | | Descriptor: | DNA (38-MER), DNA (39-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | Deposit date: | 2022-11-09 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2M8N
 
 | | HIV-1 capsid monomer structure | | Descriptor: | Capsid protein p24 | | Authors: | Deshmukh, L, Schwieters, C.D, Grishaev, A, Clore, G, Ghirlando, R. | | Deposit date: | 2013-05-24 | | Release date: | 2013-11-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Structure and Dynamics of Full-Length HIV-1 Capsid Protein in Solution.
J.Am.Chem.Soc., 135, 2013
|
|
6VUN
 
 | | Reverse Transcriptase Diabody with R83C Mutation | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Single-chain Fv | | Authors: | Chesterman, C, Arnold, E. | | Deposit date: | 2020-02-16 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Co-crystallization with diabodies: A case study for the introduction of synthetic symmetry.
Structure, 29, 2021
|
|
6VUO
 
 | |
5DX5
 
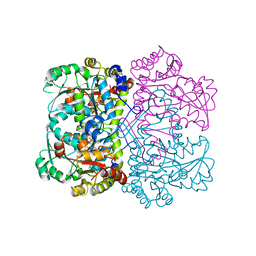 | | Crystal structure of methionine gamma-lyase from Clostridium sporogenes | | Descriptor: | CHLORIDE ION, Methionine gamma-lyase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Revtovich, S.V, Nikulin, A.D, Morozova, E.A, Anufrieva, N.V, Demidkina, T.V. | | Deposit date: | 2015-09-23 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure of methionine gamma-lyase from Clostridium sporogenes.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
1PF9
 
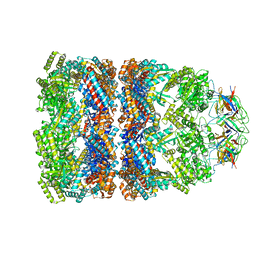 | | GroEL-GroES-ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, groEL protein, ... | | Authors: | Chaudhry, C, Farr, G.W, Todd, M.J, Rye, H.S, Brunger, A.T, Adams, P.D, Horwich, A.L, Sigler, P.B. | | Deposit date: | 2003-05-24 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.993 Å) | | Cite: | Role of the gamma-phosphate of ATP in triggering protein folding by GroEL-GroES: function, structure and energetics.
Embo J., 22, 2003
|
|
1PF3
 
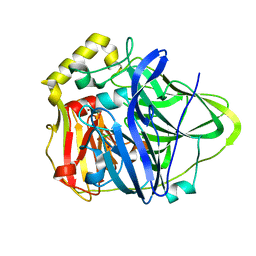 | | Crystal Structure of the M441L mutant of the multicopper oxidase CueO | | Descriptor: | Blue copper oxidase cueO, COPPER (II) ION, CU-CL-CU LINKAGE | | Authors: | Roberts, S.A, Wildner, G.F, Grass, G, Weichsel, A, Ambrus, A, Rensing, C, Montfort, W.R. | | Deposit date: | 2003-05-23 | | Release date: | 2003-06-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Labile Regulatory Copper Ion Lies Near the T1 Copper Site in the Multicopper Oxidase CueO.
J.Biol.Chem., 278, 2003
|
|
4OSR
 
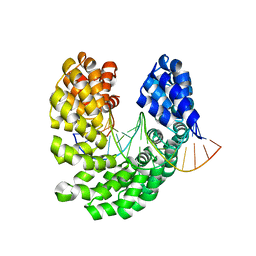 | | Crystal structure of the S505K mutant of TAL effector dHax3 | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*TP*AP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*TP*GP*TP*CP*TP*CP*TP*CP*T)-3'), Hax3 | | Authors: | Deng, D, Wu, J.P, Yan, C.Y, Pan, X.J, Yan, N. | | Deposit date: | 2014-02-13 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | Revisiting the TALE repeat
Protein Cell, 5, 2014
|
|
1PCM
 
 | |
7PQ9
 
 | | Crystal structure of Bacillus clausii pdxR at 2.8 Angstroms resolution | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Vivoli Vega, M, Isupov, M.N, Harmer, N. | | Deposit date: | 2021-09-16 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
8ENU
 
 | | Structure of the C3bB proconvertase in complex with lufaxin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3 beta chain, Complement C3b alpha' chain, ... | | Authors: | Andersen, J.F, Lei, H. | | Deposit date: | 2022-09-30 | | Release date: | 2023-08-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | A bispecific inhibitor of complement and coagulation blocks activation in complementopathy models via a novel mechanism.
Blood, 141, 2023
|
|
1PI3
 
 | | E28Q mutant Benzoylformate Decarboxylase From Pseudomonas Putida | | Descriptor: | 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Benzoylformate decarboxylase, CALCIUM ION, ... | | Authors: | Bera, A.K, Hasson, M.S. | | Deposit date: | 2003-05-29 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High resolution structure of Benzoylformate Decarboxylate E28Q mutant
To be Published
|
|
1PBY
 
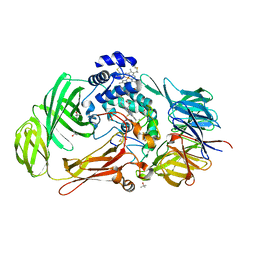 | | Structure of the Phenylhydrazine Adduct of the Quinohemoprotein Amine Dehydrogenase from Paracoccus denitrificans at 1.7 A Resolution | | Descriptor: | HEME C, TERTIARY-BUTYL ALCOHOL, quinohemoprotein amine dehydrogenase 40 kDa subunit, ... | | Authors: | Datta, S, Ikeda, T, Kano, K, Mathews, F.S. | | Deposit date: | 2003-05-15 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the phenylhydrazine adduct of the quinohemoprotein amine dehydrogenase from Paracoccus denitrificans at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2MPH
 
 | |
5E18
 
 | |
5XLB
 
 | |
8EE8
 
 | |
7NYC
 
 | | cryoEM structure of 3C9-sMAC | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Menny, A, Couves, E.C, Bubeck, D. | | Deposit date: | 2021-03-22 | | Release date: | 2021-10-06 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural basis of soluble membrane attack complex packaging for clearance.
Nat Commun, 12, 2021
|
|
7PZ2
 
 | |
8EOK
 
 | |
