2INN
 
 | | Structure of the Phenol Hydroxyalse-Regulatory Protein Complex | | Descriptor: | FE (III) ION, MOLYBDATE ION, Phenol hydroxylase component phL, ... | | Authors: | Sazinsky, M.H, Dunten, P.W, McCormick, M.S, Lippard, S.J. | | Deposit date: | 2006-10-08 | | Release date: | 2007-01-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray Structure of a Hydroxylase-Regulatory Protein Complex from a Hydrocarbon-Oxidizing Multicomponent Monooxygenase, Pseudomonas sp. OX1 Phenol Hydroxylase.
Biochemistry, 45, 2006
|
|
4DAI
 
 | | Crystal structure of B. anthracis DHPS with compound 23 | | Descriptor: | (7-amino-4,5-dioxo-1,4,5,6-tetrahydropyrimido[4,5-c]pyridazin-3-yl)acetic acid, Dihydropteroate Synthase, SULFATE ION | | Authors: | Hammoudeh, D, Lee, R.E, White, S.W. | | Deposit date: | 2012-01-12 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design of Novel Pyrimido[4,5-c]pyridazine Derivatives as Dihydropteroate Synthase Inhibitors with Increased Affinity.
Chemmedchem, 7, 2012
|
|
1UAG
 
 | | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE | | Descriptor: | SULFATE ION, UDP-N-ACETYLMURAMOYL-L-ALANINE/:D-GLUTAMATE LIGASE, URIDINE-5'-DIPHOSPHATE-N-ACETYLMURAMOYL-L-ALANINE | | Authors: | Bertrand, J, Fanchon, E, Dideberg, O. | | Deposit date: | 1997-03-13 | | Release date: | 1998-03-18 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of UDP-N-acetylmuramoyl-L-alanine:D-glutamate ligase from Escherichia coli.
EMBO J., 16, 1997
|
|
8BDY
 
 | | Crystal structure of TRIM33 alpha PHD-Bromo domain in complex with 9 | | Descriptor: | 1,3-dimethylbenzimidazol-2-one, CALCIUM ION, E3 ubiquitin-protein ligase TRIM33, ... | | Authors: | Tassone, G, Pozzi, C, Palomba, T. | | Deposit date: | 2022-10-20 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Exploiting ELIOT for Scaffold-Repurposing Opportunities: TRIM33 a Possible Novel E3 Ligase to Expand the Toolbox for PROTAC Design.
Int J Mol Sci, 23, 2022
|
|
3SGP
 
 | | Amyloid-related segment of alphaB-crystallin residues 90-100 mutant V91L | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha-crystallin B chain | | Authors: | Laganowsky, A, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2011-06-15 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4016 Å) | | Cite: | Atomic view of a toxic amyloid small oligomer.
Science, 335, 2012
|
|
1UB1
 
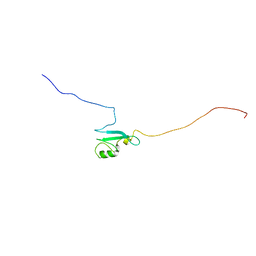 | | Solution structure of the matrix attachment region-binding domain of chicken MeCP2 | | Descriptor: | attachment region binding protein | | Authors: | Heitmann, B, Maurer, T, Weitzel, J.M, Stratling, W.H, Kalbitzer, H.R, Brunner, E, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-03-27 | | Release date: | 2003-08-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the matrix attachment region-binding domain of chicken MeCP2
EUR.J.BIOCHEM., 270, 2003
|
|
4DAT
 
 | |
8B6Y
 
 | |
1UC4
 
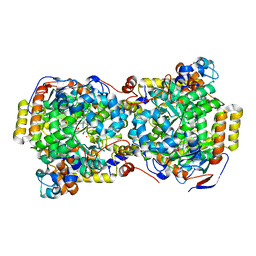 | | Structure of diol dehydratase complexed with (S)-1,2-propanediol | | Descriptor: | AMMONIUM ION, CYANOCOBALAMIN, POTASSIUM ION, ... | | Authors: | Shibata, N, Nakanishi, Y, Fukuoka, M, Yamanishi, M, Yasuoka, N, Toraya, T. | | Deposit date: | 2003-04-08 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural rationalization for the lack of stereospecificity in coenzyme B12-dependent diol dehydratase
J.BIOL.CHEM., 278, 2003
|
|
4DB5
 
 | | Crystal structure of Rabbit GITRL | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tumor necrosis factor ligand superfamily member 18 | | Authors: | Kumar, P.R, Bhosle, R, Gizzi, A, Scott Glenn, A, Chowhury, S, Hillerich, B, Seidel, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2012-01-13 | | Release date: | 2012-03-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.522 Å) | | Cite: | Crystal structure of GITRL from Oryctolagus cuniculus
to be published
|
|
8ATF
 
 | | Nucleosome-bound Ino80 ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (226-MER), DNA (227-MER), ... | | Authors: | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | Deposit date: | 2022-08-23 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
1UDD
 
 | | TenA homologue protein from P.horikoshii OT3 | | Descriptor: | transcriptional regulator | | Authors: | Itou, H, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2003-04-28 | | Release date: | 2004-06-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure analysis of PH1161 protein, a transcriptional activator TenA homologue from the hyperthermophilic archaeon Pyrococcus horikoshii.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1UE4
 
 | | Crystal structure of d(GCGAAAGC) | | Descriptor: | 5'-D(*GP*CP*GP*AP*AP*AP*GP*C)-3', MAGNESIUM ION | | Authors: | Sunami, T, Kondo, J, Hirao, I, Watanabe, K, Miura, K, Takenaka, A. | | Deposit date: | 2003-05-09 | | Release date: | 2004-03-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of d(GCGAAGC) and d(GCGAAAGC) (tetragonal form): a switching of partners of the sheared G.A pairs to form a functional G.AxA.G crossing.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
8BEG
 
 | | Structure of Ig-like domains from PrgB | | Descriptor: | MAGNESIUM ION, PrgB | | Authors: | Jarva, M, Schmitt, A, Berntsson, R.P.-A. | | Deposit date: | 2022-10-21 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural foundation for the role of enterococcal PrgB in conjugation, biofilm formation, and virulence.
Elife, 12, 2023
|
|
1U2Z
 
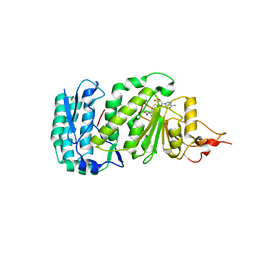 | | Crystal structure of histone K79 methyltransferase Dot1p from yeast | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Sawada, K, Yang, Z, Horton, J.R, Collins, R.E, Zhang, X, Cheng, X. | | Deposit date: | 2004-07-20 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the conserved core of the yeast Dot1p, a nucleosomal histone H3 lysine 79 methyltransferase
J.Biol.Chem., 279, 2004
|
|
8BBF
 
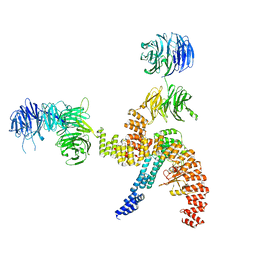 | | Structure of the IFT-A complex; IFT-A1 module | | Descriptor: | Intraflagellar transport protein 122 homolog, Intraflagellar transport protein 140 homolog, WD repeat-containing protein 19 | | Authors: | Hesketh, S.J, Mukhopadhyay, A.G, Nakamura, D, Toropova, K, Roberts, A.J. | | Deposit date: | 2022-10-12 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | IFT-A structure reveals carriages for membrane protein transport into cilia.
Cell, 185, 2022
|
|
3SGW
 
 | |
1U3G
 
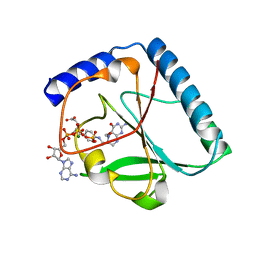 | | Structural and Functional Characterization of a 5,10-Methenyltetrahydrofolate Synthetase from Mycoplasma pneumoniae (GI: 13508087) | | Descriptor: | 5,10-Methenyltetrahydrofolate Synthetase, 5-HYDROXYMETHYLENE-6-HYDROFOLIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, S, Yakunin, A.F, Proudfoot, M, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-07-21 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of methenyltetrahydrofolate synthetase from Mycoplasma pneumoniae (GI: 13508087) at 2.2 A resolution
Proteins, 56, 2004
|
|
8BGK
 
 | |
8B4A
 
 | | Nativ complex of PqsE and RhlR with autoinducer C4-HSL | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase, FE (III) ION, N-[(3S)-2-oxotetrahydrofuran-3-yl]butanamide, ... | | Authors: | Borgert, S.R, Blankenfeldt, W. | | Deposit date: | 2022-09-20 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Moonlighting chaperone activity of the enzyme PqsE contributes to RhlR-controlled virulence of Pseudomonas aeruginosa.
Nat Commun, 13, 2022
|
|
4DDF
 
 | | Computationally Designed Self-assembling Octahedral Cage protein, O333, Crystallized in space group P4 | | Descriptor: | CHLORIDE ION, Propanediol utilization polyhedral body protein PduT, SULFATE ION | | Authors: | Sawaya, M.R, King, N.P, Sheffler, W, Baker, D, Yeates, T.O. | | Deposit date: | 2012-01-18 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Computational design of self-assembling protein nanomaterials with atomic level accuracy.
Science, 336, 2012
|
|
1U3N
 
 | | A SOD-like protein from B. subtilis, unstructured in solution, becomes ordered in the crystal: implications for function and for fibrillogenesis | | Descriptor: | Hypothetical superoxide dismutase-like protein yojM | | Authors: | Banci, L, Bertini, I, Calderone, V, Cramaro, F, Del Conte, R, Fantoni, A, Mangani, S, Quattrone, A, Viezzoli, M.S. | | Deposit date: | 2004-07-22 | | Release date: | 2005-05-03 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | A prokaryotic superoxide dismutase paralog lacking two Cu ligands: from largely unstructured in solution to ordered in the crystal.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
3SHL
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V74KL25A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Sue, G, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2011-06-16 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Redirection of internal ionizable residues by engineered cavities
To be Published
|
|
1U6H
 
 | | Vinculin head (0-258) in complex with the talin vinculin binding site 2 (849-879) | | Descriptor: | Talin, Vinculin | | Authors: | Fillingham, I, Gingras, A.R, Papagrigoriou, E, Patel, B, Emsley, J, Roberts, G.C.K, Critchley, D.R, Barsukov, I.L. | | Deposit date: | 2004-07-30 | | Release date: | 2005-01-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | A vinculin binding domain from the talin rod unfolds to form a complex with the vinculin head.
Structure, 13, 2005
|
|
8AVG
 
 | | Cryo-EM structure of mouse Elp123 with bound SAM | | Descriptor: | Elongator complex protein 1, Elongator complex protein 2, Elongator complex protein 3, ... | | Authors: | Jaciuk, M, Scherf, D, Kaszuba, K, Gaik, M, Koscielniak, A, Krutyholowa, R, Rawski, M, Indyka, P, Biela, A, Dobosz, D, Lin, T.-Y, Abbassi, N, Hammermeister, A, Chramiec-Glabik, A, Kosinski, J, Schaffrath, R, Glatt, S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Cryo-EM structure of the fully assembled Elongator complex.
Nucleic Acids Res., 51, 2023
|
|
