6UIG
 
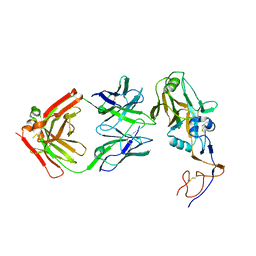 | |
3J47
 
 | | Formation of an intricate helical bundle dictates the assembly of the 26S proteasome lid | | Descriptor: | 26S proteasome regulatory subunit RPN11, 26S proteasome regulatory subunit RPN12, 26S proteasome regulatory subunit RPN3, ... | | Authors: | Estrin, E, Lopez-Blanco, J.R, Chacon, P, Martin, A. | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Formation of an Intricate Helical Bundle Dictates the Assembly of the 26S Proteasome Lid.
Structure, 21, 2013
|
|
7RVB
 
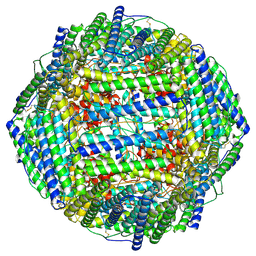 | | High resolution map of molecular chaperone Artemin | | Descriptor: | Ferritin | | Authors: | Parvate, A.D, Powell, S.M, Brookreason, J.T, Novikova, I.V, Evans, J.E. | | Deposit date: | 2021-08-18 | | Release date: | 2022-12-14 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (2.04 Å) | | Cite: | Cryo-EM structure of the diapause chaperone artemin.
Front Mol Biosci, 9, 2022
|
|
7AJU
 
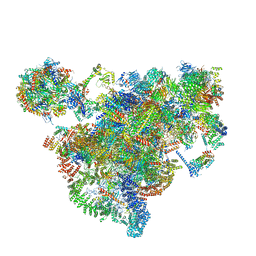 | | Cryo-EM structure of the 90S-exosome super-complex (state Post-A1-exosome) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Flemming, D, Venuta, G.L, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Maturing 90S Pre-ribosome in Association with the RNA Exosome.
Mol.Cell, 81, 2021
|
|
6P5C
 
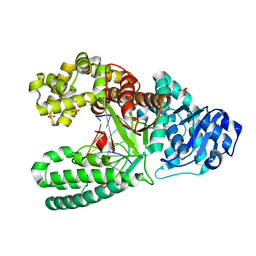 | | Bacillus Fragment DNA polymerase mutant I716M | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*AP*CP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*TP*GP*AP*TP*CP*G)-3'), DNA polymerase I, ... | | Authors: | Wu, E.Y. | | Deposit date: | 2019-05-30 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The importance of Ile716 toward the mutagenicity of 8-Oxo-2'-deoxyguanosine with Bacillus fragment DNA polymerase.
DNA Repair (Amst.), 89, 2020
|
|
5IZF
 
 | | Complex of PKA with the bisubstrate protein kinase inhibitor ARC-1408 | | Descriptor: | 6J9-ZEU-DAR-ACA-DAR-NH2, SULFATE ION, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Pflug, A, Enkvist, E, Uri, A, Engh, R.A. | | Deposit date: | 2016-03-25 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bifunctional Ligands for Inhibition of Tight-Binding Protein-Protein Interactions.
Bioconjug.Chem., 27, 2016
|
|
6PA0
 
 | | Structure of the G77A mutant in Sodium Chloride | | Descriptor: | Antibody HEAVY fragment, Antibody LIGHT fragment, DIACYL GLYCEROL, ... | | Authors: | Cuello, L.G, Guan, L. | | Deposit date: | 2019-06-11 | | Release date: | 2019-08-07 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure, function, and ion-binding properties of a K+channel stabilized in the 2,4-ion-bound configuration.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7B53
 
 | | Crystal structure of MurE from E.coli | | Descriptor: | 1,2-ETHANEDIOL, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate--2,6-diaminopimelate ligase | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
6P6A
 
 | | Structure of Mouse Importin alpha - NIT2 NLS peptide complex | | Descriptor: | Importin subunit alpha-1, Nitrogen catabolic enzyme regulatory protein | | Authors: | Bernardes, N.E, Fukuda, C.A, Silva, T.D, Oliveira, H.C, Fontes, M.R.M. | | Deposit date: | 2019-06-03 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Comparative study of the interactions between fungal transcription factor nuclear localization sequences with mammalian and fungal importin-alpha.
Sci Rep, 10, 2020
|
|
3MTH
 
 | | X-RAY CRYSTALLOGRAPHIC STUDIES ON HEXAMERIC INSULINS IN THE PRESENCE OF HELIX-STABILIZING AGENTS, THIOCYANATE, METHYLPARABEN AND PHENOL | | Descriptor: | 4-HYDROXY-BENZOIC ACID METHYL ESTER, CHLORIDE ION, METHYLPARABEN INSULIN, ... | | Authors: | Whittingham, J.L, Dodson, E.J, Moody, P.C.E, Dodson, G.G. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray crystallographic studies on hexameric insulins in the presence of helix-stabilizing agents, thiocyanate, methylparaben, and phenol.
Biochemistry, 34, 1995
|
|
1J9J
 
 | | CRYSTAL STRUCTURE ANALYSIS OF SURE PROTEIN FROM T.MARITIMA | | Descriptor: | MAGNESIUM ION, STATIONARY PHASE SURVIVAL PROTEIN, SULFATE ION | | Authors: | Suh, S.W, Lee, J.Y, Kwak, J.E, Moon, J. | | Deposit date: | 2001-05-27 | | Release date: | 2001-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and functional analysis of the SurE protein identify a novel phosphatase family.
Nat.Struct.Biol., 8, 2001
|
|
3J9X
 
 | | A Virus that Infects a Hyperthermophile Encapsidates A-Form DNA | | Descriptor: | DNA, coat protein | | Authors: | DiMaio, F, Yu, X, Rensen, E, Krupovic, M, Prangishvili, D, Egelman, E. | | Deposit date: | 2015-03-21 | | Release date: | 2015-06-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A virus that infects a hyperthermophile encapsidates A-form DNA.
Science, 348, 2015
|
|
8U1I
 
 | | Crystal structure of SyoA bound to 4-methylsyringol | | Descriptor: | 2,6-dimethoxy-4-methylphenol, Cytochrome P450, NITRATE ION, ... | | Authors: | Harlington, A.C, Shearwin, K.E, Bell, S.G, Whelan, F. | | Deposit date: | 2023-09-01 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural insights into the S-lignin O-demethylase SyoA
To Be Published
|
|
6P6Y
 
 | | Crystal structure of voltage-gated sodium channel NavAb V100C/Q150C disulfide crosslinked mutant in the activated state | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Ion transport protein | | Authors: | Wisedchaisri, G, Tonggu, L, McCord, E, Gamal El-din, T.M, Wang, L, Zheng, N, Catterall, W.A. | | Deposit date: | 2019-06-04 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Resting-State Structure and Gating Mechanism of a Voltage-Gated Sodium Channel.
Cell, 178, 2019
|
|
8U09
 
 | | Crystal structure of substrate-free SyoA | | Descriptor: | ACETATE ION, Cytochrome P450, GLYCEROL, ... | | Authors: | Harlington, A.H, Shearwin, K.E, Bell, S.G, Whelan, F. | | Deposit date: | 2023-08-28 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural insights into the S-lignin O-demethylase SyoA
To Be Published
|
|
1NAW
 
 | | ENOLPYRUVYL TRANSFERASE | | Descriptor: | CYCLOHEXYLAMMONIUM ION, UDP-N-ACETYLGLUCOSAMINE 1-CARBOXYVINYL-TRANSFERASE | | Authors: | Schoenbrunn, E, Sack, S, Eschenburg, S, Perrakis, A, Krekel, F, Amrhein, N, Mandelkow, E. | | Deposit date: | 1996-07-23 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of UDP-N-acetylglucosamine enolpyruvyltransferase, the target of the antibiotic fosfomycin.
Structure, 4, 1996
|
|
8CO0
 
 | |
4XYG
 
 | |
6P7D
 
 | | D104N S. typhimurium siroheme synthase | | Descriptor: | CHLORIDE ION, S-ADENOSYL-L-HOMOCYSTEINE, Siroheme synthase | | Authors: | Pennington, J.M, Stroupe, M.E. | | Deposit date: | 2019-06-05 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Siroheme synthase orients substrates for dehydrogenase and chelatase activities in a common active site.
Nat Commun, 11, 2020
|
|
5F7K
 
 | | Blood group antigen binding adhesin BabA of Helicobacter pylori strain 17875 in complex with Nanobody Nb-ER19 | | Descriptor: | 1,2-ETHANEDIOL, Adhesin binding fucosylated histo-blood group antigen, Nanobody Nb-ER19, ... | | Authors: | Moonens, K, Gideonsson, P, Subedi, S, Romao, E, Oscarson, S, Muyldermans, S, Boren, T, Remaut, H. | | Deposit date: | 2015-12-08 | | Release date: | 2016-01-20 | | Last modified: | 2016-12-28 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural Insights into Polymorphic ABO Glycan Binding by Helicobacter pylori.
Cell Host Microbe, 19, 2016
|
|
8U19
 
 | | Crystal structure of SyoA bound to syringol | | Descriptor: | 2,6-dimethoxyphenol, Cytochrome P450, MAGNESIUM ION, ... | | Authors: | Harlington, A.C, Shearwin, K.E, Bell, S.G, Whelan, F. | | Deposit date: | 2023-08-31 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structural insights into the S-lignin O-demethylase SyoA
To Be Published
|
|
8TCM
 
 | | Crystal Structure of modified HIV reverse transcriptase p51 domain (FPC1) with picric acid and Xanthene-1,3,6,8-tetrol bound | | Descriptor: | 9H-xanthene-1,3,6,8-tetrol, PICRIC ACID, p51 subunit | | Authors: | Pedersen, L.C, London, R.E. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal Structure of modified HIV reverse transcriptase p51 domain (FPC1) with picric acid and xanthene bound
To Be Published
|
|
6P90
 
 | | Crystal structure of PaDHDPS2-H56Q mutant | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase, CHLORIDE ION, GLYCEROL | | Authors: | Impey, R.E, Panjikar, S, Hall, C.J, Bock, L.J, Sutton, J.M, Perugini, M.A, Soares da Costa, T.P. | | Deposit date: | 2019-06-08 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of two dihydrodipicolinate synthase isoforms from Pseudomonas aeruginosa that differ in allosteric regulation.
Febs J., 287, 2020
|
|
8CIB
 
 | | Structural and functional analysis of the Pseudomonas aeruginosa PA1677 protein | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Cysteine hydrolase, ... | | Authors: | Sonnleitner, E, Brear, P, Luisi, B.F, Blasi, U. | | Deposit date: | 2023-02-09 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Catabolite repression control protein antagonist, a novel player in Pseudomonas aeruginosa carbon catabolite repression control.
Front Microbiol, 14, 2023
|
|
7RXN
 
 | |
