5SAL
 
 | | Endothiapepsin in complex with compound FU5-2 | | Descriptor: | (1Z)-1-imino-1H-isoindol-3-amine, DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Messini, N, Barthel, T, Klebe, G, Weiss, M.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-09-01 | | Last modified: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Frag4Lead: growing crystallographic fragment hits by catalog using fragment-guided template docking.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5SAT
 
 | | Endothiapepsin in complex with compound FU66-1 | | Descriptor: | Endothiapepsin, GLYCEROL, N-cyclopropyl-6-(furan-2-yl)-2-hydroxy-N-[(pyridin-2-yl)methyl]pyridine-3-carboxamide | | Authors: | Wollenhaupt, J, Metz, A, Messini, N, Barthel, T, Klebe, G, Weiss, M.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-09-01 | | Last modified: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Frag4Lead: growing crystallographic fragment hits by catalog using fragment-guided template docking.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5SAN
 
 | | Endothiapepsin in complex with compound FU5-4 | | Descriptor: | (2Z)-2-(3-amino-1H-isoindol-1-ylidene)hydrazine-1-carboximidamide, Endothiapepsin, GLYCEROL | | Authors: | Wollenhaupt, J, Metz, A, Messini, N, Barthel, T, Klebe, G, Weiss, M.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-09-01 | | Last modified: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Frag4Lead: growing crystallographic fragment hits by catalog using fragment-guided template docking.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5SAM
 
 | | Endothiapepsin in complex with compound FU5-3 | | Descriptor: | 3-amino-1H-isoindol-1-one, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Wollenhaupt, J, Metz, A, Messini, N, Barthel, T, Klebe, G, Weiss, M.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-09-01 | | Last modified: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Frag4Lead: growing crystallographic fragment hits by catalog using fragment-guided template docking.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5SAS
 
 | | Endothiapepsin in complex with compound FU290-2 | | Descriptor: | (1R)-1-(4-chlorophenyl)ethyl carbamimidothioate, BROMIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Wollenhaupt, J, Metz, A, Messini, N, Barthel, T, Klebe, G, Weiss, M.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-09-01 | | Last modified: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Frag4Lead: growing crystallographic fragment hits by catalog using fragment-guided template docking.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5SAP
 
 | | Endothiapepsin in complex with compound FU58-2 | | Descriptor: | (8S)-5-(2-aminopyrimidin-4-yl)-N-[2-(dimethylamino)ethyl]-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrazine-2-carboxamide, 1,2-ETHANEDIOL, Endothiapepsin, ... | | Authors: | Wollenhaupt, J, Metz, A, Messini, N, Barthel, T, Klebe, G, Weiss, M.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-09-01 | | Last modified: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Frag4Lead: growing crystallographic fragment hits by catalog using fragment-guided template docking.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
4DJ9
 
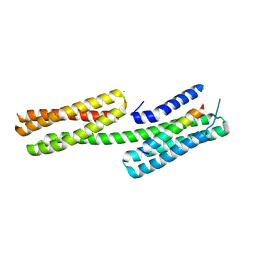 | | Human vinculin head domain Vh1 (residues 1-258) in complex with the talin vinculin binding site 50 (VBS50, residues 2078-2099) | | Descriptor: | Talin-1, Vinculin | | Authors: | Yogesha, S.D, Rangarajan, E.S, Vonrhein, C, Bricogne, G, Izard, T. | | Deposit date: | 2012-02-01 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of vinculin in complex with vinculin binding site 50 (VBS50), the integrin binding site 2 (IBS2) of talin.
Protein Sci., 21, 2012
|
|
4CL3
 
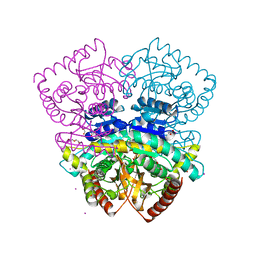 | | 1.70 A resolution structure of the malate dehydrogenase from Chloroflexus aurantiacus | | Descriptor: | ACETATE ION, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Talon, R, Madern, D, Girard, E. | | Deposit date: | 2014-01-11 | | Release date: | 2014-02-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | An Experimental Point of View on Hydration/Solvation in Halophilic Proteins.
Front.Microbiol., 5, 2014
|
|
4DR9
 
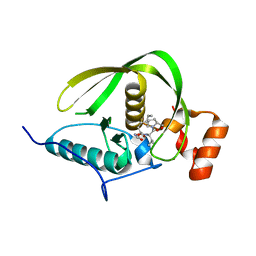 | | Crystal structure of a peptide deformylase from synechococcus elongatus in complex with actinonin | | Descriptor: | ACTINONIN, BROMIDE ION, Peptide deformylase, ... | | Authors: | Lorimer, D, Abendroth, J, Craig, T, Burgin, A, Segall, A, Rohwler, F. | | Deposit date: | 2012-02-17 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of a cyanophage-encoded peptide deformylase.
ISME J, 7, 2013
|
|
4E01
 
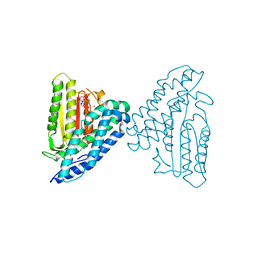 | | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/3,6-dichlorobenzo[b]thiophene-2-carboxylic acid complex with AMPPNP | | Descriptor: | 3,6-dichloro-1-benzothiophene-2-carboxylic acid, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Tso, S.C, Chuang, J.L, Gui, W.J, Wynn, R.M, Li, J, Chuang, D.T. | | Deposit date: | 2012-03-02 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structures of branched-chain alpha-ketoacid dehydrogenase kinase-inhibitor complexes
To be Published
|
|
7K7T
 
 | | Crystal structure of human MORC4 ATPase-CW in complex with AMPPNP | | Descriptor: | Isoform 3 of MORC family CW-type zinc finger protein 4, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Klein, B.J, Tencer, A.H, Kutateladze, T.G. | | Deposit date: | 2020-09-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Molecular mechanism of the MORC4 ATPase activation.
Nat Commun, 11, 2020
|
|
7KFA
 
 | | PCSK9 in complex with PCSK9i a 13mer cyclic peptide LDLR disruptor | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, CALCIUM ION, Proprotein convertase subtilisin/kexin type 9, ... | | Authors: | Chopra, R, Xu, M, Spraggon, G. | | Deposit date: | 2020-10-13 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Identification of a PCSK9-LDLR disruptor peptide with in vivo function.
Cell Chem Biol, 29, 2022
|
|
7KEV
 
 | | PCSK9 in complex with a cyclic peptide LDLR disruptor | | Descriptor: | CALCIUM ION, Proprotein convertase subtilisin/kexin type 9, Proprotein convertase subtilisin/kexin type 9 Propeptide, ... | | Authors: | Spraggon, G, Chopra, R. | | Deposit date: | 2020-10-12 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of a PCSK9-LDLR disruptor peptide with in vivo function.
Cell Chem Biol, 29, 2022
|
|
7M0V
 
 | | Crystal structure of the BRAF:MEK1 kinases in complex with AMPPNP and Cobimetinib | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, K, Gonzalez Del-Pino, G, Ha, B.H, Park, E, Eck, M.J. | | Deposit date: | 2021-03-11 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Allosteric MEK inhibitors act on BRAF/MEK complexes to block MEK activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7M0W
 
 | | Crystal structure of the BRAF:MEK1 kinases in complex with AMPPNP and Pimasertib | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, N-[(2S)-2,3-dihydroxypropyl]-3-[(2-fluoro-4-iodophenyl)amino]pyridine-4-carboxamide, ... | | Authors: | Li, K, Gonzalez Del-Pino, G, Ha, B.H, Park, E, Eck, M.J. | | Deposit date: | 2021-03-11 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Allosteric MEK inhibitors act on BRAF/MEK complexes to block MEK activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7M0U
 
 | | Crystal structure of the BRAF:MEK1 kinases in complex with AMPPNP and Binimetinib | | Descriptor: | 5-[(4-bromo-2-fluorophenyl)amino]-4-fluoro-N-(2-hydroxyethoxy)-1-methyl-1H-benzimidazole-6-carboxamide, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Li, K, Gonzalez Del-Pino, G, Ha, B.H, Park, E, Eck, M.J. | | Deposit date: | 2021-03-11 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Allosteric MEK inhibitors act on BRAF/MEK complexes to block MEK activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7M0T
 
 | | Crystal structure of the BRAF:MEK1 kinases in complex with AMPPNP and Selumetinib | | Descriptor: | 5-[(4-bromo-2-chlorophenyl)amino]-4-fluoro-N-(2-hydroxyethoxy)-1-methyl-1H-benzimidazole-6-carboxamide, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Li, K, Gonzalez Del-Pino, G, Ha, B.H, Park, E, Eck, M.J. | | Deposit date: | 2021-03-11 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Allosteric MEK inhibitors act on BRAF/MEK complexes to block MEK activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2FE6
 
 | | P450CAM from Pseudomonas putida reconstituted with manganic protoporphyrin IX | | Descriptor: | Cytochrome P450-cam, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING MN | | Authors: | von Koenig, K, Makris, T.M, Sligar, S.G, Schlichting, I. | | Deposit date: | 2005-12-15 | | Release date: | 2006-03-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The status of high-valent metal oxo complexes in the P450 cytochromes.
J.Inorg.Biochem., 100, 2006
|
|
1C8J
 
 | | CRYSTAL STRUCTURE OF CYTOCHROME P450CAM MUTANT (F87W/Y96F) | | Descriptor: | CYTOCHROME P450-CAM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Liu, Y, Jiang, F, Guo, Q, Chen, X, Jin, J, Sun, Y, Rao, Z. | | Deposit date: | 2000-05-31 | | Release date: | 2001-05-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Cytochrome P450cam mutant (F87W/Y96F)
To be Published
|
|
1OWZ
 
 | | T4 Lysozyme Cavity Mutant L99A/M102Q Bound With 4-FluoroPhenEthyl Alcohol | | Descriptor: | 4-FLUOROPHENETHYL ALCOHOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | Deposit date: | 2003-03-31 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Testing a Flexible-receptor Docking Algorithm in a Model Binding Site
J.Mol.Biol., 337, 2004
|
|
1OV5
 
 | | T4 Lysozyme Cavity Mutant L99a/M102Q Bound With 2-Allylphenol | | Descriptor: | 2-ALLYLPHENOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | Deposit date: | 2003-03-25 | | Release date: | 2004-04-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Testing a Flexible-receptor Docking Algorithm in a Model Binding Site
J.Mol.Biol., 337, 2004
|
|
1OV7
 
 | | T4 Lysozyme Cavity Mutant L99A/M102Q Bound with 2-Allyl-6-Methyl-Phenol | | Descriptor: | 2-ALLYL-6-METHYL-PHENOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | Deposit date: | 2003-03-25 | | Release date: | 2004-04-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Testing a Flexible-receptor Docking Algorithm in a Model Binding Site
J.Mol.Biol., 337, 2004
|
|
1OVJ
 
 | | T4 Lysozyme Cavity Mutant L99A/M102Q Bound with 3-Fluoro-2-Methyl_Aniline | | Descriptor: | 3-FLUORO-2-METHYL-ANILINE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | Deposit date: | 2003-03-26 | | Release date: | 2004-04-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Testing a Flexible-receptor Docking Algorithm in a Model Binding Site
J.Mol.Biol., 337, 2004
|
|
1OVK
 
 | | T4 Lysozyme Cavity Mutant L99A/M102Q Bound with N-Allyl-Aniline | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme, ... | | Authors: | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | Deposit date: | 2003-03-26 | | Release date: | 2004-04-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Testing a Flexible-receptor Docking Algorithm in a Model Binding Site
J.Mol.Biol., 337, 2004
|
|
1OVH
 
 | | T4 Lysozyme Cavity Mutant L99A/M102Q Bound With 2-Chloro-6-Methyl-Aniline | | Descriptor: | 2-CHLORO-6-METHYL-ANILINE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | Deposit date: | 2003-03-26 | | Release date: | 2004-04-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Testing a Flexible-receptor Docking Algorithm in a Model Binding Site
J.Mol.Biol., 337, 2004
|
|
