6UFP
 
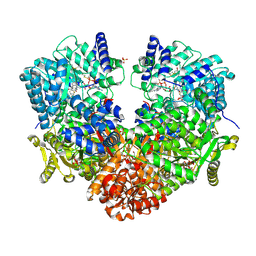 | | Structure of proline utilization A with the FAD covalently modified by L-thiazolidine-2-carboxylate and three cysteines (Cys46, Cys470, Cys638) modified to S,S-(2-HYDROXYETHYL)THIOCYSTEINE | | Descriptor: | (2S)-1,3-thiazolidine-2-carboxylic acid, Bifunctional protein PutA, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Campbell, A.C, Tanner, J.J. | | Deposit date: | 2019-09-24 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.737 Å) | | Cite: | Covalent Modification of the Flavin in Proline Dehydrogenase by Thiazolidine-2-Carboxylate.
Acs Chem.Biol., 15, 2020
|
|
9OIM
 
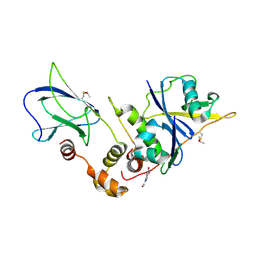 | | The von Hippel Lindau-ElonginB-ElonginC (VCB) complex with fragment 9 | | Descriptor: | 1-[5-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]-2,3-dihydroindol-1-yl]ethanone, Elongin-B, Elongin-C, ... | | Authors: | Amporndanai, K, Katinas, J.M, Chopra, A, Fesik, S.W. | | Deposit date: | 2025-05-06 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | NMR-Based Fragment Screen of the von Hippel-Lindau Elongin C&B Complex
Acs Med.Chem.Lett., 2025
|
|
9OIO
 
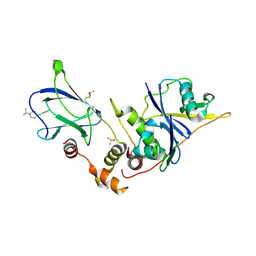 | | The von Hippel Lindau-ElonginB-ElonginC (VCB) complex with fragments 9 and 14 | | Descriptor: | 1-[(2-fluorophenyl)methyl]-4-(propan-2-yl)piperazine, 1-[6-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]-2,3-dihydroindol-1-yl]ethanone, DIMETHYL SULFOXIDE, ... | | Authors: | Amporndanai, K, Katinas, J.M, Chopra, A, Fesik, S.W. | | Deposit date: | 2025-05-06 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | NMR-Based Fragment Screen of the von Hippel-Lindau Elongin C&B Complex
Acs Med.Chem.Lett., 2025
|
|
9OIN
 
 | | The von Hippel Lindau-ElonginB-ElonginC (VCB) complex with fragment 13 | | Descriptor: | 1-(propan-2-yl)-4-[(pyridin-2-yl)methyl]piperazine, Elongin-B, Elongin-C, ... | | Authors: | Amporndanai, K, Katinas, J.M, Chopra, A, Fesik, S.W. | | Deposit date: | 2025-05-06 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | NMR-Based Fragment Screen of the von Hippel-Lindau Elongin C&B Complex
Acs Med.Chem.Lett., 2025
|
|
9OIQ
 
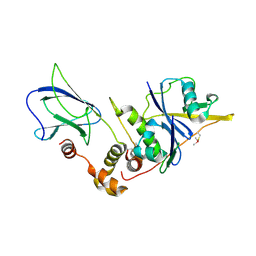 | | The von Hippel Lindau-ElonginB-ElonginC (VCB) complex with fragment 15 | | Descriptor: | 2-(5-amino-1H-1,3-benzimidazol-1-yl)ethan-1-ol, Elongin-B, Elongin-C, ... | | Authors: | Amporndanai, K, Katinas, J.M, Chopra, A, Fesik, S.W. | | Deposit date: | 2025-05-06 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | NMR-Based Fragment Screen of the von Hippel-Lindau Elongin C&B Complex
Acs Med.Chem.Lett., 2025
|
|
6VZ9
 
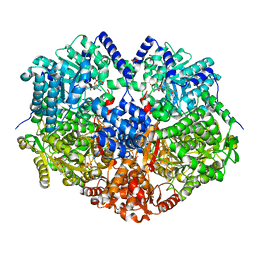 | |
6QWQ
 
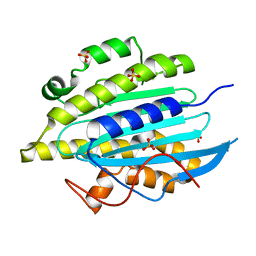 | | Structure of gtPebB | | Descriptor: | Ferredoxin bilin reductase plastid, SULFATE ION | | Authors: | Sommerkamp, J.A, Hofmann, E. | | Deposit date: | 2019-03-06 | | Release date: | 2019-08-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the first eukaryotic bilin reductaseGtPEBB reveals a flipped binding mode of dihydrobiliverdin.
J.Biol.Chem., 294, 2019
|
|
7JJQ
 
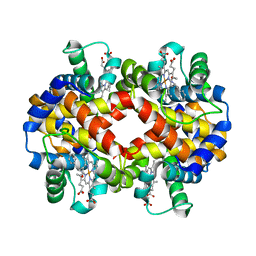 | | Human Hemoglobin in Complex with Nitrosoamphetamine | | Descriptor: | (2R)-N-hydroxy-1-phenylpropan-2-amine, GLYCEROL, Hemoglobin subunit alpha, ... | | Authors: | Powell, S.M, Thomas, L.M, Richter-Addo, G.B. | | Deposit date: | 2020-07-27 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The nitrosoamphetamine metabolite is accommodated in the active site of human hemoglobin: Spectroscopy and crystal structure.
J.Inorg.Biochem., 213, 2020
|
|
1C3Q
 
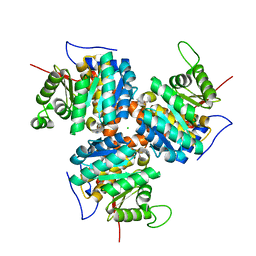 | | CRYSTAL STRUCTURE OF NATIVE THIAZOLE KINASE IN THE MONOCLINIC FORM | | Descriptor: | 2-(4-METHYL-THIAZOL-5-YL)-ETHANOL, CHLORIDE ION, Hydroxyethylthiazole kinase | | Authors: | Campobasso, N, Mathews, I.I, Begley, T.P, Ealick, S.E. | | Deposit date: | 1999-07-28 | | Release date: | 1999-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of 4-methyl-5-beta-hydroxyethylthiazole kinase from Bacillus subtilis at 1.5 A resolution.
Biochemistry, 39, 2000
|
|
2THI
 
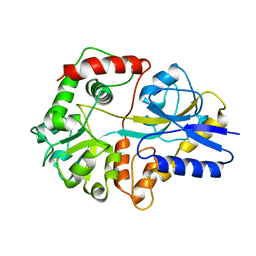 | |
4HKT
 
 | |
4LUP
 
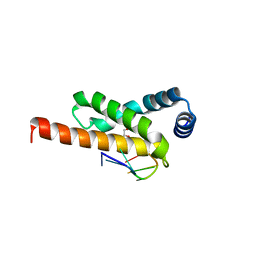 | | Crystal structure of the complex formed by region of E. coli sigmaE bound to its -10 element non template strand | | Descriptor: | 1,2-ETHANEDIOL, RNA polymerase sigma factor, region 2 of sigmaE of E. coli | | Authors: | Campagne, S, Marsh, M.E, Vorholt, J.A.V, Allain, F.H.-T, Capitani, G. | | Deposit date: | 2013-07-25 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis for -10 promoter element melting by environmentally induced sigma factors.
Nat.Struct.Mol.Biol., 21, 2014
|
|
3HQC
 
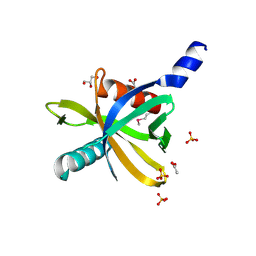 | | Crystal structure of Phosphotyrosine-binding domain from the Human Tensin-like C1 domain-containing phosphatase (TENC1) | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Sampathkumar, P, Romero, R, Wasserman, S, Do, J, Dickey, M, Bain, K, Gheyi, T, Klemke, R, Atwell, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-06-05 | | Release date: | 2009-07-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Phosphotyrosine-binding domain from the Human Tensin-like C1 domain-containing phosphatase (TENC1)
To be Published
|
|
6MEL
 
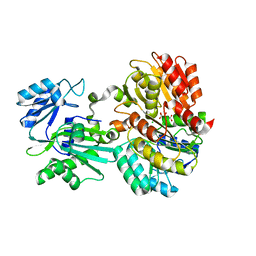 | | Succinyl-CoA synthase from Campylobacter jejuni | | Descriptor: | CHLORIDE ION, CITRIC ACID, Succinate--CoA ligase [ADP-forming] subunit beta, ... | | Authors: | Osipiuk, J, Maltseva, N, Jedrzejczak, R, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-06 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Succinyl-CoA synthase from Campylobacter jejuni
to be published
|
|
3HWJ
 
 | | Crystal structure of the second PHR domain of Mouse Myc-binding protein 2 (MYCBP-2) | | Descriptor: | DIMETHYL SULFOXIDE, E3 ubiquitin-protein ligase MYCBP2 | | Authors: | Sampathkumar, P, Ozyurt, S.A, Wasserman, S.R, Miller, S.A, Bain, K.T, Rutter, M.E, Gheyi, T, Klemke, R.L, Atwell, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-06-17 | | Release date: | 2009-07-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of PHR domains from Mus musculus Phr1 (Mycbp2) explain the loss-of-function mutation (Gly1092-->Glu) of the C. elegans ortholog RPM-1.
J.Mol.Biol., 397, 2010
|
|
9CRN
 
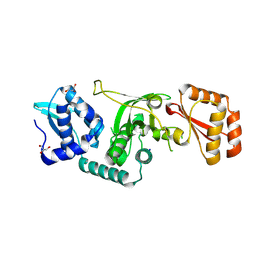 | |
4IFQ
 
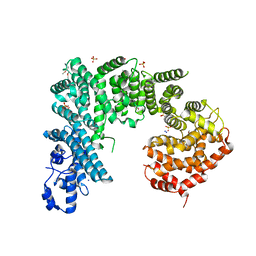 | | Crystal structure of Saccharomyces cerevisiae NUP192, residues 2 to 960 [ScNup192(2-960)] | | Descriptor: | IODIDE ION, Nucleoporin NUP192, SULFATE ION | | Authors: | Sampathkumar, P, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXstals) | | Deposit date: | 2012-12-14 | | Release date: | 2013-02-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure, dynamics, evolution, and function of a major scaffold component in the nuclear pore complex.
Structure, 21, 2013
|
|
4JJM
 
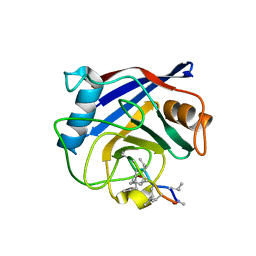 | | Structure of a cyclophilin from Citrus sinensis (CsCyp) in complex with cyclosporin A | | Descriptor: | Peptidyl-prolyl cis-trans isomerase, cyclosporin A | | Authors: | Campos, B.M, Ambrosio, A.L.B, Souza, T.A.C.B, Barbosa, J.A.R.G, Benedetti, C.E. | | Deposit date: | 2013-03-08 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | A redox 2-cys mechanism regulates the catalytic activity of divergent cyclophilins.
Plant Physiol., 162, 2013
|
|
4IPG
 
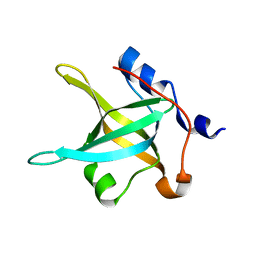 | | Structure of the N-terminal domain of RPA70, E7R, E100R mutant | | Descriptor: | Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Vangamudi, B, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-01-09 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Surface Reengineering of RPA70N Enables Cocrystallization with an Inhibitor of the Replication Protein A Interaction Motif of ATR Interacting Protein.
Biochemistry, 52, 2013
|
|
4IPH
 
 | | Structure of N-terminal domain of RPA70 in complex with VU079104 inhibitor | | Descriptor: | Replication protein A 70 kDa DNA-binding subunit, ~{N}-(2,3-dimethylphenyl)-7-oxidanylidene-12-sulfanylidene-5,11-dithia-1,8-diazatricyclo[7.3.0.0^{2,6}]dodeca-2(6),3,9-triene-10-carboxamide | | Authors: | Feldkamp, M.D, Frank, A.O, Vangamudi, B, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-01-09 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Surface Reengineering of RPA70N Enables Cocrystallization with an Inhibitor of the Replication Protein A Interaction Motif of ATR Interacting Protein.
Biochemistry, 52, 2013
|
|
4IPY
 
 | | HIV capsid C-terminal domain | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein p24 | | Authors: | Lampel, A, Yaniv, O, Berger, O, Bachrach, E, Gazit, E, Frolow, F. | | Deposit date: | 2013-01-10 | | Release date: | 2013-10-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A triclinic crystal structure of the carboxy-terminal domain of HIV-1 capsid protein with four molecules in the asymmetric unit reveals a novel packing interface.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4JWT
 
 | |
4JP2
 
 | | Crystal Structure of TT0495 protein from Thermus thermophilus HB8 | | Descriptor: | 2-deoxy-D-gluconate 3-dehydrogenase | | Authors: | Pampa, K.J, Lokanath, N.K, Kunishima, N, Ravishnkar Rai, V. | | Deposit date: | 2013-03-19 | | Release date: | 2014-03-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | The first crystal structure of NAD-dependent 3-dehydro-2-deoxy-D-gluconate dehydrogenase from Thermus thermophilus HB8
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7BAJ
 
 | |
7BAK
 
 | |
