7OHV
 
 | |
7OHX
 
 | |
7OHP
 
 | |
7OHR
 
 | | Nog1-TAP associated immature ribosomal particle population E from S. cerevisiae | | Descriptor: | 25S rRNA, 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Milkereit, P, Poell, G. | | Deposit date: | 2021-05-11 | | Release date: | 2021-11-10 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.72 Å) | | Cite: | Analysis of subunit folding contribution of three yeast large ribosomal subunit proteins required for stabilisation and processing of intermediate nuclear rRNA precursors.
Plos One, 16, 2021
|
|
4NO0
 
 | | Crystal structure of non-phosphorylated form of RQA_V phosphopeptide bound to HLA-A2 in complex with LILRB1 | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Mohammed, F, Stones, D.H, Willcox, B.E. | | Deposit date: | 2013-11-19 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The antigenic identity of human class I MHC phosphopeptides is critically dependent upon phosphorylation status.
Oncotarget, 8, 2017
|
|
8V84
 
 | | 60S ribosome biogenesis intermediate (Dbp10 catalytic structure - Overall map) | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | Authors: | Cruz, V.E, Weirich, C.S, Peddada, N, Erzberger, J.P. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The DEAD-box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60S ribosome biogenesis.
Nat Commun, 15, 2024
|
|
8V83
 
 | | 60S ribosome biogenesis intermediate (Dbp10 pre-catalytic structure - Overall map) | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | Authors: | Cruz, V.E, Weirich, C.S, Peddada, N, Erzberger, J.P. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | The DEAD-box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60S ribosome biogenesis.
Nat Commun, 15, 2024
|
|
8V87
 
 | | 60S ribosome biogenesis intermediate (Dbp10 post-catalytic structure - Overall map) | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 25S ribosomal RNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Cruz, V.E, Weirich, C.S, Peddada, N, Erzberger, J.P. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | The DEAD-box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60S ribosome biogenesis.
Nat Commun, 15, 2024
|
|
8RTB
 
 | |
8RT7
 
 | |
1J8H
 
 | | Crystal Structure of a Complex of a Human alpha/beta-T cell Receptor, Influenza HA Antigen Peptide, and MHC Class II Molecule, HLA-DR4 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ HA1 PEPTIDE CHAIN, ... | | Authors: | Hennecke, J, Wiley, D.C. | | Deposit date: | 2001-05-21 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a complex of the human alpha/beta T cell receptor (TCR) HA1.7, influenza hemagglutinin peptide, and major histocompatibility complex class II molecule, HLA-DR4 (DRA*0101 and DRB1*0401): insight into TCR cross-restriction and alloreactivity.
J.Exp.Med., 195, 2002
|
|
8E5T
 
 | | Yeast co-transcriptional Noc1-Noc2 RNP assembly checkpoint intermediate | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 60S ribosomal protein L13-A, ... | | Authors: | Sanghai, Z.A, Piwowarczyk, R, Vanden Broeck, A, Klinge, S. | | Deposit date: | 2022-08-22 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | A co-transcriptional ribosome assembly checkpoint controls nascent large ribosomal subunit maturation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8EUP
 
 | | Ytm1 associated 60S nascent ribosome State 1A | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8EV3
 
 | | Ytm1 associated 60S nascent ribosome (-Fkbp39) State 1B | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8ESQ
 
 | | Ytm1 associated nascent 60S ribosome State 2 | | Descriptor: | 25S rRNA (cytosine-C(5))-methyltransferase nop2, 60S ribosomal protein L13, 60S ribosomal protein L14, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-14 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8EUY
 
 | | Ytm1 associated nascent 60S ribosome (-fkbp39) State 1A | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8ETH
 
 | | Ytm1 associated 60S nascent ribosome State 1B | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-17 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8ESR
 
 | | Ytm1 associated nascent 60S ribosome (-fkbp39) State 2 | | Descriptor: | 25S rRNA (cytosine-C(5))-methyltransferase nop2, 60S ribosomal protein L13, 60S ribosomal protein L14, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-14 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8ETI
 
 | | Fkbp39 associated 60S nascent ribosome State 1 | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-17 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8ETG
 
 | | Fkbp39 associated 60S nascent ribosome State 3 | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-17 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8EIZ
 
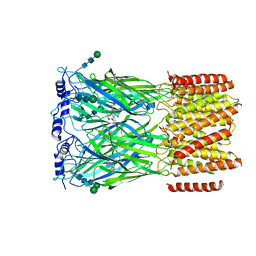 | | Cryo-EM structure of squid sensory receptor CRB1 | | Descriptor: | N-benzyl-2-(2,6-dimethylanilino)-N,N-diethyl-2-oxoethan-1-aminium, Squid sensory receptor CRB1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kang, G, Kim, J.J, Allard, C.A.H, Valencia-Montoya, W.A, van Giesen, L, Kilian, P.B, Bai, X, Bellono, N.W, Hibbs, R.E. | | Deposit date: | 2022-09-15 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Sensory specializations drive octopus and squid behaviour.
Nature, 616, 2023
|
|
8RT5
 
 | |
8RT6
 
 | |
8RT8
 
 | |
8RT4
 
 | |
