5F46
 
 | | Crystal structure of an aminoglycoside acetyltransferase meta-AAC0020 from an uncultured soil metagenomic sample, apoenzyme form | | Descriptor: | CHLORIDE ION, aminoglycoside acetyltransferase meta-AAC0020 | | Authors: | Xu, Z, Skarina, T, Stogios, P.J, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-03 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Functional Survey of Environmental Aminoglycoside Acetyltransferases Reveals Functionality of Resistance Enzymes.
ACS Infect Dis, 3, 2017
|
|
5F48
 
 | | Crystal structure of an aminoglycoside acetyltransferase meta-AAC0020 from an uncultured soil metagenomic sample in complex with coenzyme A | | Descriptor: | CHLORIDE ION, COENZYME A, MAGNESIUM ION, ... | | Authors: | Xu, Z, Skarina, T, Stogios, P.J, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-03 | | Release date: | 2015-12-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Functional Survey of Environmental Aminoglycoside Acetyltransferases Reveals Functionality of Resistance Enzymes.
ACS Infect Dis, 3, 2017
|
|
5F47
 
 | | Crystal structure of an aminoglycoside acetyltransferase meta-AAC0020 from an uncultured soil metagenomic sample in complex with trehalose | | Descriptor: | CALCIUM ION, CHLORIDE ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, ... | | Authors: | Xu, Z, Skarina, T, Wawrzak, Z, Stogios, P.J, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-03 | | Release date: | 2015-12-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | Structural and Functional Survey of Environmental Aminoglycoside Acetyltransferases Reveals Functionality of Resistance Enzymes.
ACS Infect Dis, 3, 2017
|
|
2R1I
 
 | |
2ZPA
 
 | | Crystal Structure of tRNA(Met) Cytidine Acetyltransferase | | Descriptor: | ACETYL COENZYME *A, ADENOSINE-5'-DIPHOSPHATE, SULFATE ION, ... | | Authors: | Chimnaronk, S, Manita, T, Yao, M, Tanaka, I. | | Deposit date: | 2008-07-08 | | Release date: | 2009-04-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | RNA helicase module in an acetyltransferase that modifies a specific tRNA anticodon
Embo J., 28, 2009
|
|
6D6Y
 
 | |
7KYQ
 
 | |
7KYS
 
 | | Crystal structure of human BCCIP beta (Native2) | | Descriptor: | 1,2-ETHANEDIOL, BRCA2 and CDKN1A-interacting protein, DI(HYDROXYETHYL)ETHER | | Authors: | Choi, W.S, Liu, B, Shen, Z, Yang, W. | | Deposit date: | 2020-12-08 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of human BCCIP and implications for binding and modification of partner proteins.
Protein Sci., 30, 2021
|
|
4ONU
 
 | |
4ORF
 
 | |
4OLL
 
 | |
6B3A
 
 | |
6B3B
 
 | | AprA Methyltransferase 1 - GNAT in complex with Mn2+ , SAM, and Malonate | | Descriptor: | AprA Methyltransferase 1, GLYCEROL, MALONATE ION, ... | | Authors: | Skiba, M.A, Smith, J.L. | | Deposit date: | 2017-09-21 | | Release date: | 2017-11-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Mononuclear Iron-Dependent Methyltransferase Catalyzes Initial Steps in Assembly of the Apratoxin A Polyketide Starter Unit.
ACS Chem. Biol., 12, 2017
|
|
6B39
 
 | | AprA Methyltransferase 1 - GNAT in complex with SAH | | Descriptor: | AprA Methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION | | Authors: | Skiba, M.A, Smith, J.L. | | Deposit date: | 2017-09-21 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | A Mononuclear Iron-Dependent Methyltransferase Catalyzes Initial Steps in Assembly of the Apratoxin A Polyketide Starter Unit.
ACS Chem. Biol., 12, 2017
|
|
8ZT3
 
 | |
8ZT4
 
 | | Complex structure of N-acetyltransferase SbzI and carrier protein SbzG in the biosynthesis of altemicidin | | Descriptor: | BICARBONATE ION, CHLORIDE ION, Carrier protein,GNAT family transferase, ... | | Authors: | Zhu, Y, Mori, T, Abe, I. | | Deposit date: | 2024-06-06 | | Release date: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-function analysis of carrier protein-dependent 2-sulfamoylacetyl transferase in the biosynthesis of altemicidin.
Nat Commun, 15, 2024
|
|
4JWP
 
 | |
4J3G
 
 | |
1CM0
 
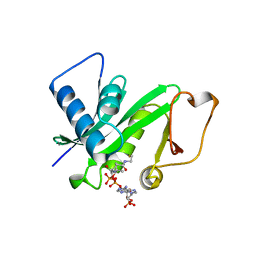 | | CRYSTAL STRUCTURE OF THE PCAF/COENZYME-A COMPLEX | | Descriptor: | COENZYME A, P300/CBP ASSOCIATING FACTOR | | Authors: | Clements, A, Rojas, J.R, Trievel, R.C, Wang, L, Berger, S.L, Marmorstein, R. | | Deposit date: | 1999-05-12 | | Release date: | 1999-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the histone acetyltransferase domain of the human PCAF transcriptional regulator bound to coenzyme A.
EMBO J., 18, 1999
|
|
9ISQ
 
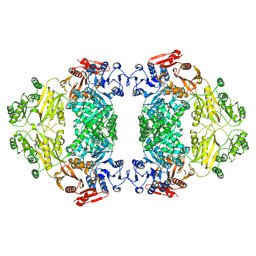 | | Apo-state E.coli PatZ | | Descriptor: | Protein acetyltransferase | | Authors: | Park, J.B, Roh, S.H. | | Deposit date: | 2024-07-18 | | Release date: | 2025-06-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Structural basis of the catalytic and allosteric mechanism of bacterial acetyltransferase PatZ.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9IT0
 
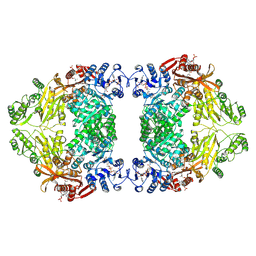 | | Liganded-state E.coli PatZ | | Descriptor: | ACETYL COENZYME *A, PHOSPHATE ION, Protein acetyltransferase | | Authors: | Park, J.B, Roh, S.H. | | Deposit date: | 2024-07-19 | | Release date: | 2025-06-04 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (1.99 Å) | | Cite: | Structural basis of the catalytic and allosteric mechanism of bacterial acetyltransferase PatZ.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ISB
 
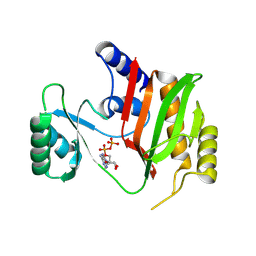 | | Ligand bound AGD of enzyme | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Protein acetyltransferase | | Authors: | Park, J.B, Roh, S.H. | | Deposit date: | 2024-07-17 | | Release date: | 2025-06-04 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural basis of the catalytic and allosteric mechanism of bacterial acetyltransferase PatZ.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
3F0A
 
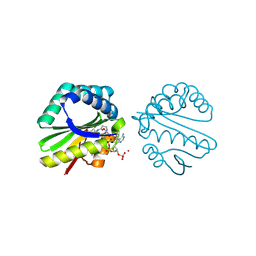 | | Structure of a putative n-acetyltransferase (ta0374) in complex with acetyl-coa from thermoplasma acidophilum | | Descriptor: | ACETYL COENZYME *A, CHLORIDE ION, N-ACETYLTRANSFERASE, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-24 | | Release date: | 2008-11-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the novel PaiA N-acetyltransferase from Thermoplasma acidophilum involved in the negative control of sporulation and degradative enzyme production.
Proteins, 79, 2011
|
|
3FIX
 
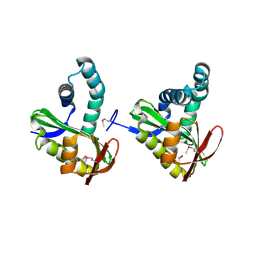 | | Crystal structure of a putative n-acetyltransferase (ta0374) from thermoplasma acidophilum | | Descriptor: | 1,2-ETHANEDIOL, N-ACETYLTRANSFERASE | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-12 | | Release date: | 2009-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the novel PaiA N-acetyltransferase from Thermoplasma acidophilum involved in the negative control of sporulation and degradative enzyme production.
Proteins, 79, 2011
|
|
3K9U
 
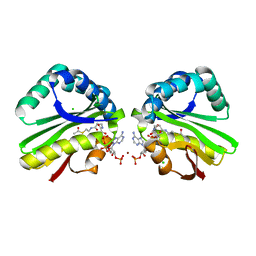 | | Crystal structure of paia acetyltransferase (ta0374) from thermoplasma acidophilum | | Descriptor: | ACETYL COENZYME *A, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-16 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the novel PaiA N-acetyltransferase from Thermoplasma acidophilum involved in the negative control of sporulation and degradative enzyme production.
Proteins, 79, 2011
|
|
