6U3D
 
 | |
6UCG
 
 | |
6X6F
 
 | | The structure of Pf6r from the filamentous phage Pf6 of Pseudomonas aeruginosa PA01 | | Descriptor: | NITRATE ION, Pf6r | | Authors: | Michie, K.A, Norrian, P, Duggin, I.G, McDougald, D, Rice, S.A. | | Deposit date: | 2020-05-28 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.735 Å) | | Cite: | The Repressor C Protein, Pf4r, Controls Superinfection of Pseudomonas aeruginosa PAO1 by the Pf4 Filamentous Phage and Regulates Host Gene Expression.
Viruses, 13, 2021
|
|
6TVZ
 
 | | Structure of a psychrophilic CCA-adding enzyme crystallized in the XtalController device | | Descriptor: | ACETATE ION, CCA-adding enzyme, GLYCEROL, ... | | Authors: | de Wijn, R, Rollet, K, Coudray, L, Hennig, O, Betat, H, Moerl, M, Lorber, B, Sauter, C. | | Deposit date: | 2020-01-10 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Monitoring the Production of High Diffraction-Quality Crystals of Two Enzymes in Real Time Using In Situ Dynamic Light Scattering
Crystals, 2020
|
|
6U9H
 
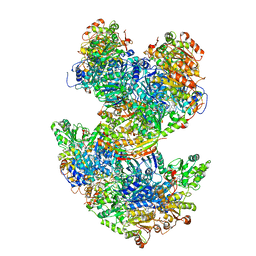 | | Arabidopsis thaliana acetohydroxyacid synthase complex | | Descriptor: | Acetolactate synthase small subunit 2, chloroplastic, Acetolactate synthase, ... | | Authors: | Guddat, L.W, Low, Y.S, Rao, Z. | | Deposit date: | 2019-09-08 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of fungal and plant acetohydroxyacid synthases.
Nature, 586, 2020
|
|
6A97
 
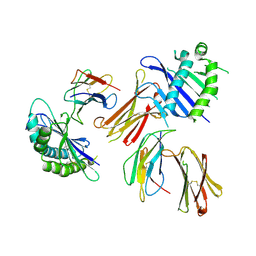 | | Crystal structure of MHC-like MILL2 | | Descriptor: | Beta-2-microglobulin, MHC I-like leukocyte 2 long form, SULFATE ION | | Authors: | Kajikawa, M, Ose, T, Maenaka, K. | | Deposit date: | 2018-07-11 | | Release date: | 2018-12-05 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | Structure of MHC class I-like MILL2 reveals heparan-sulfate binding and interdomain flexibility.
Nat Commun, 9, 2018
|
|
6UIH
 
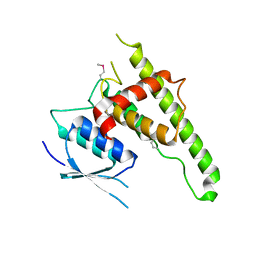 | |
6ULR
 
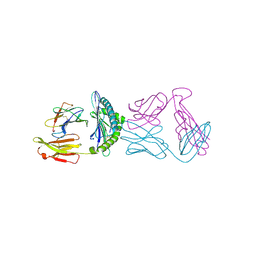 | |
6X8B
 
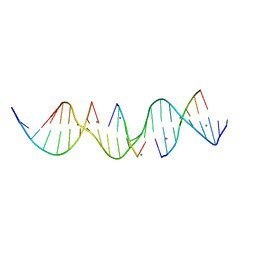 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J6 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*AP*AP*GP*AP*CP*TP*TP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*A)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-06-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6X8C
 
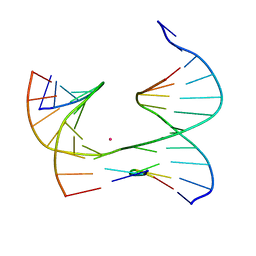 | | Self-assembly of a 3D DNA crystal lattice (4x5 junction version) containing the J1 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*TP*GP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-06-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.098 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6XDV
 
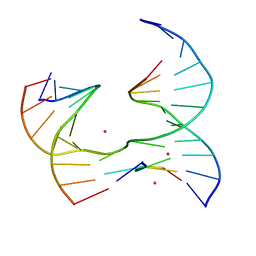 | | Self-assembly of a 3D DNA crystal lattice (4x5 junction version) containing the J3 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*TP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*GP*CP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-06-11 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6XEM
 
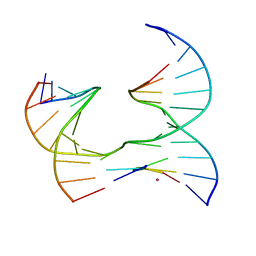 | | Self-assembly of a 3D DNA crystal lattice (4x5 junction version) containing the J16 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*TP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*GP*CP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-06-12 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.055 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6XFC
 
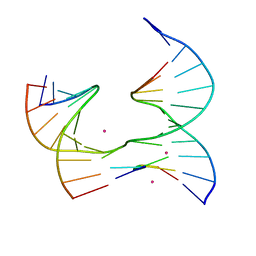 | | Self-assembly of a 3D DNA crystal lattice (4x5 junction version) containing the J19 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*GP*CP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-06-15 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.756 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6XFE
 
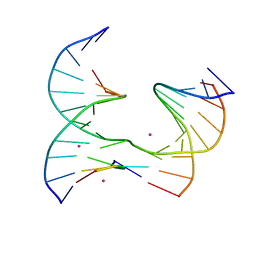 | | Self-assembly of a 3D DNA crystal lattice (4x5 junction version) containing the J21 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*CP*GP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-06-15 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6XFZ
 
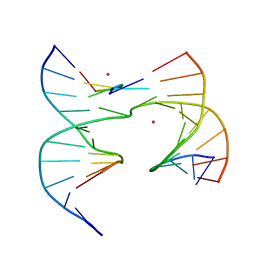 | | Self-assembly of a 3D DNA crystal lattice (4x5 junction version) containing the J29 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*AP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*CP*TP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-06-16 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.113 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6XDX
 
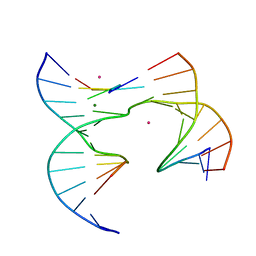 | | Self-assembly of a 3D DNA crystal lattice (4x5 junction version) containing the J6 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*AP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*AP*TP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-06-11 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6XG0
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 junction version) containing the J31 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*GP*GP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-06-16 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.178 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6XDW
 
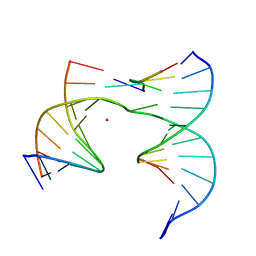 | | Self-assembly of a 3D DNA crystal lattice (4x5 junction version) containing the J5 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*CP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*CP*GP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-06-11 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.148 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6XGO
 
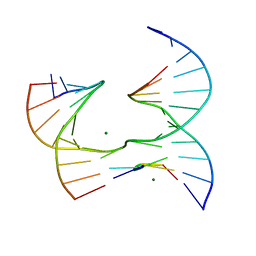 | | Self-assembly of a 3D DNA crystal lattice (4x5 junction version) containing the J34 immobile Holliday junction | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*AP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*GP*TP*GP*TP*CP*TP*GP*C)-3'), DNA (5'-D(P*AP*CP*TP*CP*CP*AP*CP*TP*CP*A)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-06-17 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6XGM
 
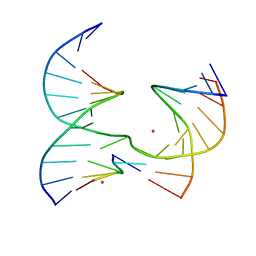 | | Self-assembly of a 3D DNA crystal lattice (4x5 junction version) containing the J25 immobile Holliday junction | | Descriptor: | COBALT (II) ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*CP*CP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-06-17 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6XDY
 
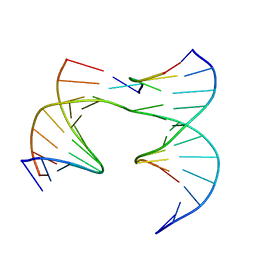 | | Self-assembly of a 3D DNA crystal lattice (4x5 junction version) containing the J7 immobile Holliday junction | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*AP*GP*GP*TP*CP*TP*GP*C)-3'), DNA (5'-D(P*AP*CP*GP*TP*CP*AP*CP*TP*CP*A)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-06-11 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6XEI
 
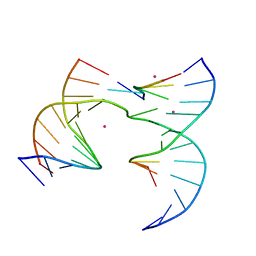 | | Self-assembly of a 3D DNA crystal lattice (4x5 junction version) containing the J9 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*TP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*CP*CP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-06-12 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.049 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6XO8
 
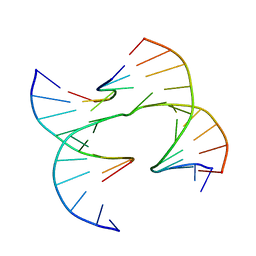 | | Self-assembly of a 3D DNA crystal lattice (4x6 junction version) containing the J31 immobile Holliday junction | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*GP*GP*TP*CP*TP*GP*C)-3'), DNA (5'-D(P*CP*GP*GP*CP*AP*CP*TP*CP*A)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-06 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (4.195 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6XEL
 
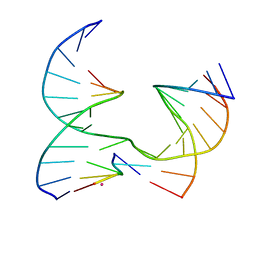 | | Self-assembly of a 3D DNA crystal lattice (4x5 junction version) containing the J15 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*TP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*CP*CP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-06-12 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6XO7
 
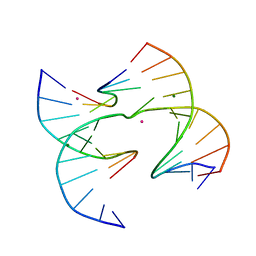 | | Self-assembly of a 3D DNA crystal lattice (4x6 junction version) containing the J30 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*CP*GP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-06 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.105 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
